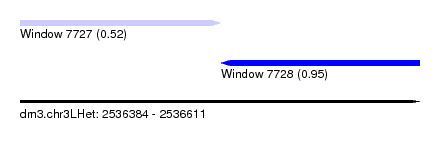
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 2,536,384 – 2,536,611 |
| Length | 227 |
| Max. P | 0.950889 |

| Location | 2,536,384 – 2,536,498 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.47 |
| Shannon entropy | 0.55751 |
| G+C content | 0.53662 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 2536384 114 + 2555491 UUCGGCAUGGUAAAUGGCAUCGGAAACGGAGGUUGGUUGCUUCCUCUCCACUGGUACUGCGACCAGACGCCUGUUUCCCUAUGGUUUUCAACUGUCUGGGGCUCUUCUCUCUCC--- (((((.(((((....(((...((((((((.(.((((((((..((........))....)))))))).)..)))))))).....)))....))))))))))..............--- ( -31.50, z-score = 0.33, R) >droSim1.chrU 5056725 117 + 15797150 CUUGGCAUGGCAAAUGGCAUUGAAAACGGAGGAGGAUUGUUUCCUCCCCAAUGGUACUGCGACCAAAUGCCUGUUUCUCUGUGAGCUUCCAAAAUCGGUGGCUCUUCCUUCUUCUGA ....(((.((.((((((((((......((.((((((.....))))))))..((((......)))))))))).)))))).)))(((((.((......)).)))))............. ( -38.40, z-score = -2.09, R) >droSec1.super_165 59835 114 + 68140 UUUGGCAUAGUAAAUGGCAUCGGAAACGGAGGUUGGUCGUUUCCUCCCCACCGGUACUGCGGCCAACCGCCUGUUUCCCUAUGGGUUCCACUUGGUUGGGACUCCUCUUUCUCC--- ..((.(((.....))).))..((((((((.((((((((((..((........))....))))))))))..))))))))....((((((((......))))))))..........--- ( -43.40, z-score = -2.15, R) >droYak2.chr2h_random 2814463 114 - 3774259 CUUUGCAU---AAACGGCAUUGGAAACGGAGGAGGGUUAGUCCCUCCCCACUGAUACUGGGACCAAAUGCCCGUCUCCCGGUGAGCCUCCAACGUUUGGGGCUUCUCUUUCUCCUGA ...(((..---.....)))..(((...((((..((((..(((((..............))))).....))))..)))).(..(((((.(((.....))))))))..)....)))... ( -37.04, z-score = -0.16, R) >droEre2.scaffold_4929 21783330 99 + 26641161 ------------------UUCGGCAUUGGAAACGGACUGCUUCCUCCCCACUGGUACUGCGACCAGACGCGUGUUUCCCUGUGGGUUUUCAACACCUGGGACUCUUCCUUCUCCUGA ------------------...((..(((((((((((.....)))...((((.((.((((((......)))).))...)).))))))))))))..)).((((....))))........ ( -26.90, z-score = -0.07, R) >consensus CUUGGCAU_G_AAAUGGCAUCGGAAACGGAGGAGGGUUGCUUCCUCCCCACUGGUACUGCGACCAAACGCCUGUUUCCCUGUGGGUUUCCAACGUCUGGGGCUCUUCUUUCUCCUGA .....................((((((((.....((((((..((........))....))))))......))))))))....((((((((......))))))))............. (-19.24 = -19.84 + 0.60)

| Location | 2,536,498 – 2,536,611 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.22 |
| Shannon entropy | 0.55117 |
| G+C content | 0.45532 |
| Mean single sequence MFE | -28.29 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 2536498 113 - 2555491 AUAGCUUGCAGGGAAAAUAACUUGGUGGCCUUGGCAGAAACCAAUUCAACUCAGAAAGCACAGGAGUCAACGGAGUCAACAGAUAAGGCUACUGAGGAAGUUCGAAAACAAGU ...(((((....(((.....((..(((((((((.(.(((.....))).((((.....((......)).....)))).....).)))))))))..))....))).....))))) ( -27.20, z-score = -0.99, R) >droSim1.chrU 5056842 113 - 15797150 AUAGCCUGUAGGGAGGAAAAGUUAGUUGCCUUAGAAGGUGGCGGCUCAGAACCUAAAACUCAAGGGUCGGUAGAGUCGCGAGAUAAGGCUAUUAAAGAAACUGAGAAACAGGA ....(((((.....((.....(((((.((((((.....(.(((((((.((.(((........))).))....))))))).)..)))))).))))).....)).....))))). ( -33.00, z-score = -1.98, R) >droSec1.super_165 59949 107 - 68140 AUAGCUUGCAGGGAAAGGAACCUGUUAGCCUUGGCGGAA------UCAGAUCGGAAAGCACAGGACUCAACGGAGUCAAAAGAUAAGGCUACUGAGGAAGUUCAGAAACAAGA ...(((.(((((........))))).)))((((.(.(((------(....((((..(((....(((((....)))))..........))).))))....)))).)...)))). ( -28.74, z-score = -1.62, R) >droYak2.chr2h_random 2814577 113 + 3774259 AUAGUUUGUAGAGAGGAGAAGUUAACGGCUGUAGAAAGUGUCGGCUUAGAACCCAAAACGCAGAGGUCGUUGGAGUCGCGAGACAAAGCCAUUAAAGAAACUCAGAAACAGGA ...((((.(.(((.............(((..........)))(((((.....((((...((....))..)))).(((....))).)))))..........)))).)))).... ( -24.20, z-score = -0.22, R) >consensus AUAGCUUGCAGGGAAAAGAACUUAGUGGCCUUAGAAGAAG_CGGCUCAGAACCGAAAACACAGGAGUCAACGGAGUCAAGAGAUAAGGCUACUAAAGAAACUCAGAAACAAGA ....(((((...(((.....((((((((((((.........((((((................)))))).....(((....)))))))))))))))....)))....))))). (-19.08 = -19.52 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:01 2011