
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 2,505,513 – 2,505,564 |
| Length | 51 |
| Max. P | 0.807485 |

| Location | 2,505,513 – 2,505,564 |
|---|---|
| Length | 51 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 64.56 |
| Shannon entropy | 0.59113 |
| G+C content | 0.57579 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -7.19 |
| Energy contribution | -6.67 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 2505513 51 - 2555491 -AGGGCCGUCAUAGCGAGGCGU--------UGUAGCCGACGCCUAGCGCAGUUUCGGCGU -...((((..((.(((((((((--------((....))))))))..))).))..)))).. ( -24.00, z-score = -2.46, R) >droSec1.super_63 175113 51 + 208916 -AGGACCGUUAUAGCGAGGCGU--------UGUAGCCGAUGUCUAGCGCAGUUUCGACGU -.....((..((.(((((((((--------((....))))))))..))).))..)).... ( -13.30, z-score = 0.03, R) >droSim1.chrU 11959274 51 + 15797150 -AGAACCGUUAUAGCGAGGCAU--------UGUUGCCGAUGCAUAGCGCAGUUUCGGCGU -....(((..((.(((..((((--------((....))))))....))).))..)))... ( -16.30, z-score = -1.24, R) >droAna3.scaffold_12905 7308 60 + 569613 AAGUAUCGUUGUAGCGAGGCGACAGCGUUGUGUCGCUGUCGCCGAUCGAGGUUCUGUAGU .....((((....))))((((((((((......))))))))))................. ( -24.60, z-score = -3.07, R) >droMoj3.scaffold_6416 46196 58 + 86987 AUGUCCCGUUGUAGCUGGGCGACAGCGUU-UGCAGCUGUCGCUGAGUGUGGUUCGGUGG- .....(((.....(((.(((((((((...-....))))))))).)))......)))...- ( -26.00, z-score = -3.04, R) >consensus _AGGACCGUUAUAGCGAGGCGU________UGUAGCCGACGCCUAGCGCAGUUUCGGCGU .............((.((((((................)))))).))............. ( -7.19 = -6.67 + -0.52)

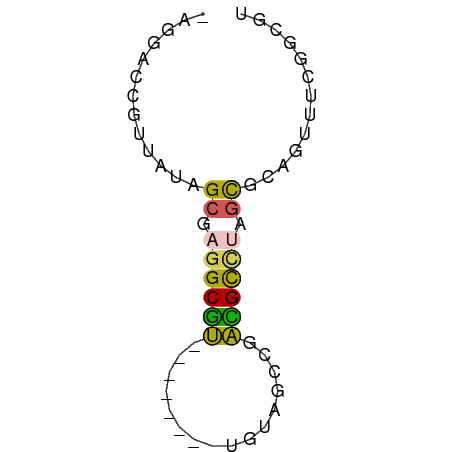

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:59 2011