| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 2,491,205 – 2,491,270 |
| Length | 65 |
| Max. P | 0.938275 |

| Location | 2,491,205 – 2,491,270 |
|---|---|
| Length | 65 |
| Sequences | 5 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 70.96 |
| Shannon entropy | 0.50491 |
| G+C content | 0.45540 |
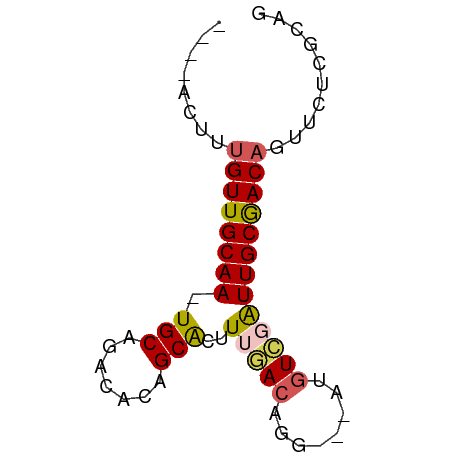
| Mean single sequence MFE | -16.03 |
| Consensus MFE | -11.54 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
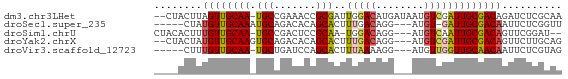
>dm3.chr3LHet 2491205 65 - 2555491 --CUACUUAGUUGCAA-UGCCGAAACCGCGAUUGGACAUGAUAAUGUCGAUUGCGACAGAUCUCGCAA --.......(((((((-(((.......)).....(((((....))))).))))))))........... ( -15.70, z-score = -1.14, R) >droSec1.super_235 11272 59 - 24486 -----CUAUGUUGCAAAUGCAGACACAGCACUUUGACAGG---AUGU-GAUUGCGACAAUUCUCGGUU -----...((((((((.((((..(..............).---.)))-).)))))))).......... ( -12.34, z-score = -0.06, R) >droSim1.chrU 5567486 61 - 15797150 CUACACUUUGUUGCAA-UGCCGACUCCGCAA-UGGACAGG---AUGUCAAUUGCGACAGUUCGGAU-- .......(((((((((-(...((((((....-......))---).))).)))))))))).......-- ( -17.40, z-score = -1.53, R) >droYak2.chrX 20704572 63 - 21770863 --CUACUAUGUUGCAAGUGCAGACACAGCACUUUGACAGG---AUGUCGAUUGCGACAGUUCUUGCAG --......((((((((((((.......)))).(((((...---..))))))))))))).......... ( -18.50, z-score = -1.17, R) >droVir3.scaffold_12723 361266 59 - 5802038 -----CUUUGUUGCAA-UGCUGAUCCAGCACUUUAAAAGG---AUGUUGGUUGCAACAAUUCUCGUAG -----..(((((((((-.......(((((((((....)))---.)))))))))))))))......... ( -16.21, z-score = -2.17, R) >consensus ____ACUUUGUUGCAA_UGCAGACACAGCACUUUGACAGG___AUGUCGAUUGCGACAGUUCUCGCAG ........((((((((.(((.......)))..((((((......)))))))))))))).......... (-11.54 = -11.82 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:59 2011