
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 2,445,663 – 2,445,739 |
| Length | 76 |
| Max. P | 0.999119 |

| Location | 2,445,663 – 2,445,739 |
|---|---|
| Length | 76 |
| Sequences | 8 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 63.25 |
| Shannon entropy | 0.75884 |
| G+C content | 0.59916 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
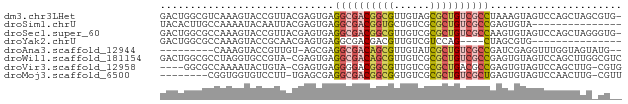
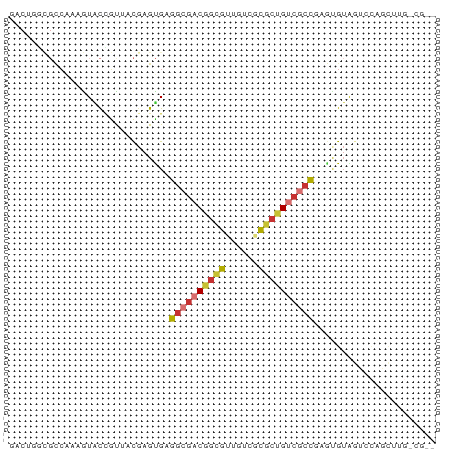
>dm3.chr3LHet 2445663 76 + 2555491 GACUGGCGUCAAAGUACCGUUACGAGUGAGGCGACGGCGUUGUAGCGCUGUCGCCUAAAGUAGUCCAGCUAGCGUG- (.(((((.(((..(((....)))...)))(((((((((((....)))))))))))............))))))...- ( -31.40, z-score = -2.30, R) >droSim1.chrU 10271674 62 - 15797150 UACACUUGCCAAAAUACAAUUACGAGUGAGGCGACGGUGCUGUCGCGCUGUCGCCGAGUGUA--------------- (((((((.......(((........))).(((((((((((....))))))))))))))))))--------------- ( -28.50, z-score = -4.12, R) >droSec1.super_60 150380 76 + 163194 GACUGGCGCCAAAGUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAGGGUG- ..(((((..((..(((....)))...)).(((((((((((....)))))))))))............)))))....- ( -30.40, z-score = -1.29, R) >droYak2.chrU 19814955 58 - 28119190 GACUGGCGCCAAAGUACCGCAACGAGUGAGGCGACGACGUUGUCGUCCAG----CUAGCGUG--------------- (.((((((((....(((((...)).))).)))((((((...))))))..)----)))))...--------------- ( -19.40, z-score = -0.58, R) >droAna3.scaffold_12944 226763 65 - 272910 ---------CAAAGUACCGUUGU-AGCGAGGCGACAGCGUUGUAUCGCUGUCGCCGAUCGAGGUUUGGUAGUAUG-- ---------...(.(((((....-..(((((((((((((......))))))))))..))).....))))).)...-- ( -28.40, z-score = -4.36, R) >droWil1.scaffold_181154 1830346 76 + 4610121 GACUGGCGCCUAGGUGCCGUA-CGAGUGAGGCGACAGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCUUGGCGUC ....((((((.((.((...((-((.....(((((((((((....)))))))))))...))))...)).)).)))))) ( -38.30, z-score = -2.68, R) >droVir3.scaffold_12958 1410437 71 - 3547706 ----GGCGCCAAAAUACUGUA-CGAGUGAGGGGACGGCGUUGUCGCGCUGACGCCGAGUGUAGUCCAGCUUG-CGUG ----.((((......((((((-(......(....)((((((........))))))..))))))).......)-))). ( -20.82, z-score = 1.34, R) >droMoj3.scaffold_6500 31239524 67 + 32352404 --------CGGUGGUGUCCUU-UGAGCGAGGCGACGGCGGUGUCGCGCUGUCGCUGAGUGUAGUCCAACUUG-CGUU --------..(..(((..((.-...((..((((((((((......))))))))))..))..))..))..)..-)... ( -24.70, z-score = -0.36, R) >consensus GACUGGCGCCAAAGUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCUUG_CG__ .............................((((((((((......))))))))))...................... (-15.97 = -16.62 + 0.66)

| Location | 2,445,663 – 2,445,739 |
|---|---|
| Length | 76 |
| Sequences | 8 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 63.25 |
| Shannon entropy | 0.75884 |
| G+C content | 0.59916 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -9.86 |
| Energy contribution | -11.00 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
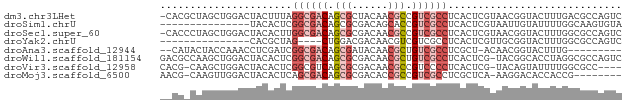
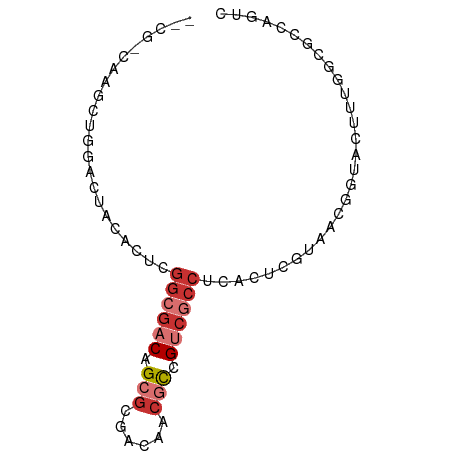
>dm3.chr3LHet 2445663 76 - 2555491 -CACGCUAGCUGGACUACUUUAGGCGACAGCGCUACAACGCCGUCGCCUCACUCGUAACGGUACUUUGACGCCAGUC -.......(((((........(((((((.(((......))).))))))).....(((....))).......))))). ( -23.00, z-score = -1.03, R) >droSim1.chrU 10271674 62 + 15797150 ---------------UACACUCGGCGACAGCGCGACAGCACCGUCGCCUCACUCGUAAUUGUAUUUUGGCAAGUGUA ---------------(((((..((((((.(.((....)).).))))))..........((((......))))))))) ( -20.60, z-score = -2.01, R) >droSec1.super_60 150380 76 - 163194 -CACCCUAGCUGGACUACACUUGGCGACAGCGCGACAACGCCGUCGCCUCACUCGUAACGGUACUUUGGCGCCAGUC -.......((..((.(((....((((((.(((......))).)))))).....((...))))).))..))....... ( -23.10, z-score = -0.55, R) >droYak2.chrU 19814955 58 + 28119190 ---------------CACGCUAG----CUGGACGACAACGUCGUCGCCUCACUCGUUGCGGUACUUUGGCGCCAGUC ---------------..((((((----...((((((...))))))(((.((.....)).)))...))))))...... ( -16.60, z-score = 0.16, R) >droAna3.scaffold_12944 226763 65 + 272910 --CAUACUACCAAACCUCGAUCGGCGACAGCGAUACAACGCUGUCGCCUCGCU-ACAACGGUACUUUG--------- --.....((((......(((..((((((((((......)))))))))))))..-.....)))).....--------- ( -24.52, z-score = -5.92, R) >droWil1.scaffold_181154 1830346 76 - 4610121 GACGCCAAGCUGGACUACACUCGGCGACAGCGCGACAACGCUGUCGCCUCACUCG-UACGGCACCUAGGCGCCAGUC (.((((..((((.((.......((((((((((......))))))))))......)-).)))).....)))).).... ( -30.82, z-score = -2.34, R) >droVir3.scaffold_12958 1410437 71 + 3547706 CACG-CAAGCUGGACUACACUCGGCGUCAGCGCGACAACGCCGUCCCCUCACUCG-UACAGUAUUUUGGCGCC---- ....-...((..((.(((.....(((....)))(((......)))..........-....))).))..))...---- ( -15.10, z-score = 1.31, R) >droMoj3.scaffold_6500 31239524 67 - 32352404 AACG-CAAGUUGGACUACACUCAGCGACAGCGCGACACCGCCGUCGCCUCGCUCA-AAGGACACCACCG-------- (((.-...)))((.........(((((..(.(((((......)))))))))))..-..((...)).)).-------- ( -17.30, z-score = -0.85, R) >consensus __CG_CAAGCUGGACUACACUCGGCGACAGCGCGACAACGCCGUCGCCUCACUCGUAACGGUACUUUGGCGCCAGUC ......................((((((.(((......))).))))))............................. ( -9.86 = -11.00 + 1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:56 2011