| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 2,378,635 – 2,378,745 |
| Length | 110 |
| Max. P | 0.959458 |

| Location | 2,378,635 – 2,378,745 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Shannon entropy | 0.37663 |
| G+C content | 0.45056 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -22.04 |
| Energy contribution | -21.93 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
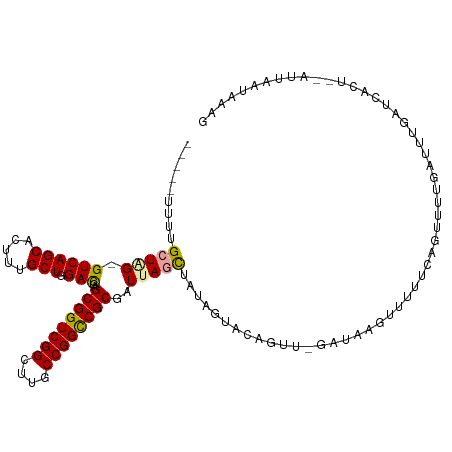
>dm3.chr3LHet 2378635 110 - 2555491 -------UUGCUGGUGUCAGCACUUUGCUGGAUGAGCGGUCGGCUUGCCGGUCGCGGUUAGCUCUAGUACAGUUGGAUAAGUUUUUCAGUUUUGCUAUGAUCGCUGAUUCAAUAAAG -------.(((((....)))))(((((.(((((.(((((((((((.((((....)))).)))).(((((.(..((((.......))))..).))))).))))))).))))).))))) ( -41.30, z-score = -3.32, R) >droEre2.scaffold_4845 1110261 112 - 22589142 GCUGUUUUUGGUAGAGUCAGCA-UUUGCUGGACGAGCGGUCGGCUUGCCGGCCGCGAUUAGCUAUAGUACAGUU-GAUAAGUUUUUCAGUUUAGUUUUGAUCAC---CUUAAUAAAG ..((((...(((.((.((((.(-((.((((((...((((((((....)))))))).(((((((.......))))-)))......))))))..))).))))))))---)..))))... ( -34.50, z-score = -2.20, R) >droAna3.scaffold_13261 33980 111 + 284793 -GCUGUUUUGCUAG-GUCAGCACUUCGCUAGACGAGCGGUCGGUUUGCCGGCCGCGCUAAGUUAAAGUACAGUUA---AGAGUUUUCAGUUUUGAUUUGA-CAUUCUACUAAUAAAG -(((((...(((((-((((((.....))).)))..((((((((....)))))))).)).)))......)))))..---(((((..((((.......))))-.))))).......... ( -32.60, z-score = -1.43, R) >consensus _____UUUUGCUAG_GUCAGCACUUUGCUGGACGAGCGGUCGGCUUGCCGGCCGCGAUUAGCUAUAGUACAGUU_GAUAAGUUUUUCAGUUUUGAUUUGAUCACU__AUUAAUAAAG .........(((((.((((((.....))).)))..((((((((....))))))))..)))))....................................................... (-22.04 = -21.93 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:53 2011