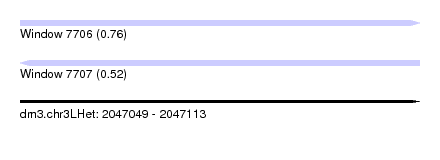
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 2,047,049 – 2,047,113 |
| Length | 64 |
| Max. P | 0.764200 |

| Location | 2,047,049 – 2,047,113 |
|---|---|
| Length | 64 |
| Sequences | 5 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 66.92 |
| Shannon entropy | 0.60348 |
| G+C content | 0.43858 |
| Mean single sequence MFE | -14.52 |
| Consensus MFE | -7.58 |
| Energy contribution | -8.86 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
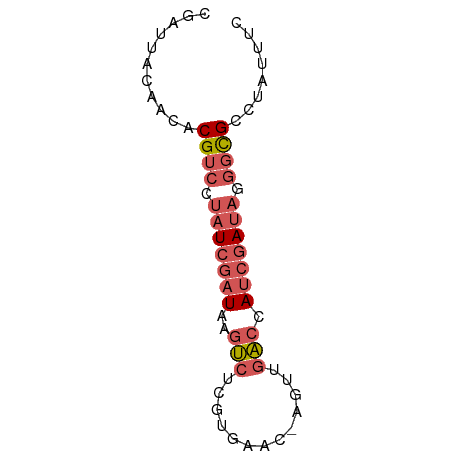
>dm3.chr3LHet 2047049 64 + 2555491 GAGAUAGGCGCCCUAUCGAUGGUCAACU-GUUCACGAGACUUAUCGAUAGGACGUGUUGUAAUCG ((....((((((((((((((((((....-........))).))))))))))..)))))....)). ( -21.50, z-score = -2.44, R) >droSim1.chrU 10617183 64 - 15797150 GAAAUAGGCGCCCUAUCGAUGGUCAACU-GUUCCCGAGACUUAUCGAUAUGACGAGUUGUAAUCG ...(((((...)))))((((...(((((-(((..(((......)))....))).)))))..)))) ( -12.70, z-score = -0.07, R) >droSec1.super_74 143438 63 + 162789 AAAAUAGGCGCCCUAUCGAUGGUCAACG-GUUCACGAGACUUAUCGAUAUGACGAGUU-UAAUCG ....(((((..(.(((((((((((..((-.....)).))).)))))))).)....)))-)).... ( -13.80, z-score = -0.51, R) >droYak2.chrU 27751680 65 - 28119190 GAAAUGAGCGCCUUAUCCAUGGUUAACUUGUUUACGAUACUAAUCGAUAGGACGGUUUGUAUUUG .(((..((.(((........)))...))..)))..(((((.(((((......))))).))))).. ( -12.40, z-score = -0.80, R) >droAna3.scaffold_13469 689026 56 + 871550 --------CAGUCUCUCGUUAGACAUUAGAGUUUAAGGUGUGAUC-AUUGGUCGUGCUGGAAUUU --------..((((......))))....((((((..((..((((.-....))))..)).)))))) ( -12.20, z-score = -1.00, R) >consensus GAAAUAGGCGCCCUAUCGAUGGUCAACU_GUUCACGAGACUUAUCGAUAGGACGAGUUGUAAUCG ........((.(((((((((((((.............)))).))))))))).))........... ( -7.58 = -8.86 + 1.28)



| Location | 2,047,049 – 2,047,113 |
|---|---|
| Length | 64 |
| Sequences | 5 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 66.92 |
| Shannon entropy | 0.60348 |
| G+C content | 0.43858 |
| Mean single sequence MFE | -11.92 |
| Consensus MFE | -5.68 |
| Energy contribution | -7.00 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 2047049 64 - 2555491 CGAUUACAACACGUCCUAUCGAUAAGUCUCGUGAAC-AGUUGACCAUCGAUAGGGCGCCUAUCUC ...........((((((((((((..(((..(....)-....))).))))))))))))........ ( -20.40, z-score = -3.11, R) >droSim1.chrU 10617183 64 + 15797150 CGAUUACAACUCGUCAUAUCGAUAAGUCUCGGGAAC-AGUUGACCAUCGAUAGGGCGCCUAUUUC ...........((((.(((((((..(((..(....)-....))).))))))).))))........ ( -14.00, z-score = -0.58, R) >droSec1.super_74 143438 63 - 162789 CGAUUA-AACUCGUCAUAUCGAUAAGUCUCGUGAAC-CGUUGACCAUCGAUAGGGCGCCUAUUUU ......-....((((.(((((((..(((.((.....-))..))).))))))).))))........ ( -14.60, z-score = -0.97, R) >droYak2.chrU 27751680 65 + 28119190 CAAAUACAAACCGUCCUAUCGAUUAGUAUCGUAAACAAGUUAACCAUGGAUAAGGCGCUCAUUUC ...........((((.((((.((...((.(........).))...)).)))).))))........ ( -7.50, z-score = -0.20, R) >droAna3.scaffold_13469 689026 56 - 871550 AAAUUCCAGCACGACCAAU-GAUCACACCUUAAACUCUAAUGUCUAACGAGAGACUG-------- ...................-.....................((((......))))..-------- ( -3.10, z-score = 0.53, R) >consensus CGAUUACAACACGUCCUAUCGAUAAGUCUCGUGAAC_AGUUGACCAUCGAUAGGGCGCCUAUUUC ...........((((.(((((((..(((.............))).))))))).))))........ ( -5.68 = -7.00 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:44 2011