
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,985,432 – 3,985,526 |
| Length | 94 |
| Max. P | 0.656476 |

| Location | 3,985,432 – 3,985,526 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Shannon entropy | 0.38772 |
| G+C content | 0.36777 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.51 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
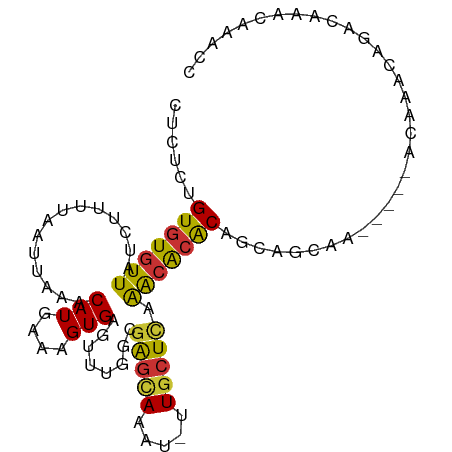
>dm3.chr2L 3985432 94 - 23011544 UUCUCUGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAGUUUGGCGAGCAAAU-UUGCUCAAACACACUGCAGCAA-----ACAAACAGACAAACAAACC ...(((((.((((....((((........))))((((.((((((((((....)-)))).))))).))))......)-----))).))))).......... ( -20.00, z-score = -1.09, R) >droSim1.chr2L 3945977 99 - 22036055 CUCUCUGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAGUUUGGCGAGCAAAU-UUGCUCAAACACACUGCAGCAAACAAGACAAACAGACAAACAAACC ...(((((.((((....((((........))))((((.((((((((((....)-)))).))))).))))...........)))).))))).......... ( -20.40, z-score = -0.84, R) >droSec1.super_5 2088284 99 - 5866729 CUCUCUGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAGUUUGGCGAGCAAAU-UUGCUCAAACACACUGCAGCAAACAAGACAAACAGACAAACAAACC ...(((((.((((....((((........))))((((.((((((((((....)-)))).))))).))))...........)))).))))).......... ( -20.40, z-score = -0.84, R) >droYak2.chr2L 4001347 94 - 22324452 CUCUCUGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAGUUUGGCGAGCAAAU-UUGCUCAAACACACAGCAGCAA-----ACAAACAGACAAACAAACC ..(((((((((((..............(((....)))........(((((...-.))))).))))))))).))...-----................... ( -22.00, z-score = -1.85, R) >droEre2.scaffold_4929 4043936 94 - 26641161 CUCUCUGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAGUUUGGCGAGCAAAU-UUGCUCAAACACACAGCAGCAA-----ACAAACAGACAAACAAACC ..(((((((((((..............(((....)))........(((((...-.))))).))))))))).))...-----................... ( -22.00, z-score = -1.85, R) >droPer1.super_1 6793163 86 + 10282868 GUGUGUGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAG-UUGGCGAGCAAAU-UUGCUCAAACACACAGCAG--------ACAAACAAACAAACC---- ((.((((((((((..............(((....)))..-.....(((((...-.))))).)))))))).)).--------)).............---- ( -23.50, z-score = -2.01, R) >droWil1.scaffold_180703 1945160 85 - 3946847 GUAUGCGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAG-UUGGCGAGUAAAU-UUGCUUAAAAACGCACAAAC-------ACAAGCAAACAUC------ ((.(((.((((((...(((((........)))))(((.(-((...(((((...-.)))))...))).))).)))-------))).))).))...------ ( -19.90, z-score = -1.72, R) >droVir3.scaffold_12963 10923009 96 + 20206255 GCGUGUGUAUGUUAUCUUUUAAUUAAACAUAAAUGUGAG-UUGCCUGCCAAGUGAUUUUUGGGCGCACAACAAUAA---UAACAACCACAACAACAUGCG ((((((.((((((............))))))..((((.(-(((....(((((.....)))))..(.....).....---...))))))))...)))))). ( -19.40, z-score = 0.14, R) >consensus CUCUCUGUGUGUUAUCUUUUAAUUAAACAUGAAAGUGAGUUUGGCGAGCAAAU_UUGCUCAAACACACAGCAGCAA_____ACAAACAGACAAACAAACC ......(((((((..............(((....)))........(((((.....))))).)))))))................................ (-14.62 = -14.51 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:06 2011