| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,996,805 – 1,996,901 |
| Length | 96 |
| Max. P | 0.964546 |

| Location | 1,996,805 – 1,996,901 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 57.29 |
| Shannon entropy | 0.61638 |
| G+C content | 0.43019 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -12.95 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
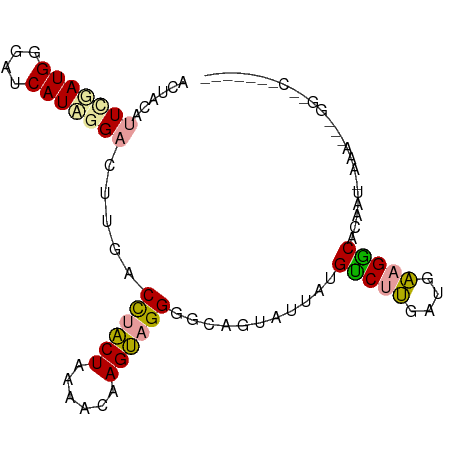
>dm3.chr3LHet 1996805 96 + 2555491 CUUAAUUUCGAUGGGAUCAUUGGCCUCGACUUGCUAGUGCAAGCUGGGGCAUCGCUUUGCCUGGUCUCCGGCCAGCUUAAAUGGGGCACGGAAGUU ...((((((...(((((((...((((((.(((((....))))).))))))...((...)).)))))))(((((..(......).))).)))))))) ( -32.40, z-score = -0.48, R) >droPer1.super_109 48952 96 - 183219 ACUACAUUUUAUGAGAUCAUAGGACUUGACCUACUAAAACAAGUAGGGGCAGUAUUAUGUCUUGAUGAAGACACAAUAAAAAUAGGUGCAACUUCG .........((((....))))(.(((((.((((((......)))))).....((((.((((((....)))))).))))....))))).)....... ( -21.10, z-score = -2.03, R) >dp4.Unknown_group_9 5316 76 + 53872 ACUACAUUCGAUGGGAUCAUAUGACUUGACCUACUAAAACAAGUAGGGGCAGUAUUAUGUCUUGAUGAAGGCACAA-------------------- ......((((.(((((.((((..(((...((((((......))))))...)))..))))))))).)))).......-------------------- ( -17.60, z-score = -1.58, R) >consensus ACUACAUUCGAUGGGAUCAUAGGACUUGACCUACUAAAACAAGUAGGGGCAGUAUUAUGUCUUGAUGAAGGCACAAU_AAA___GG__C_______ ......(((((((....))))))).....((((((......))))))...........(((((....)))))........................ (-12.95 = -12.73 + -0.21)

| Location | 1,996,805 – 1,996,901 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 57.29 |
| Shannon entropy | 0.61638 |
| G+C content | 0.43019 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -9.02 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 1996805 96 - 2555491 AACUUCCGUGCCCCAUUUAAGCUGGCCGGAGACCAGGCAAAGCGAUGCCCCAGCUUGCACUAGCAAGUCGAGGCCAAUGAUCCCAUCGAAAUUAAG ...(((.(((...((((...(((.((((....)..)))..)))...(((.(.((((((....)))))).).))).))))....))).)))...... ( -24.40, z-score = -0.59, R) >droPer1.super_109 48952 96 + 183219 CGAAGUUGCACCUAUUUUUAUUGUGUCUUCAUCAAGACAUAAUACUGCCCCUACUUGUUUUAGUAGGUCAAGUCCUAUGAUCUCAUAAAAUGUAGU ..................(((((((((((....)))))))))))((((.((((((......))))))........((((....))))....)))). ( -21.70, z-score = -2.83, R) >dp4.Unknown_group_9 5316 76 - 53872 --------------------UUGUGCCUUCAUCAAGACAUAAUACUGCCCCUACUUGUUUUAGUAGGUCAAGUCAUAUGAUCCCAUCGAAUGUAGU --------------------...(((.(((.....((((((..(((...((((((......))))))...)))..)))).)).....))).))).. ( -15.00, z-score = -1.61, R) >consensus _______G__CC___UUU_AUUGUGCCUUCAUCAAGACAUAAUACUGCCCCUACUUGUUUUAGUAGGUCAAGUCCUAUGAUCCCAUCGAAUGUAGU ....................(((((((((....))))))))).......((((((......))))))............................. ( -9.02 = -9.80 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:40 2011