| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,848,829 – 1,848,927 |
| Length | 98 |
| Max. P | 0.994487 |

| Location | 1,848,829 – 1,848,924 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.42020 |
| G+C content | 0.51689 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -25.76 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
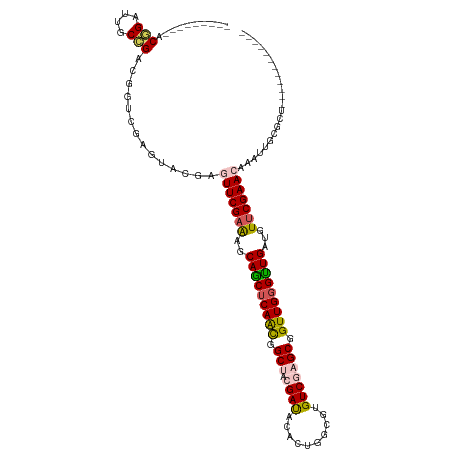
>dm3.chr3LHet 1848829 95 + 2555491 ------------ACAGAUUGCCGACGGUCGAGUACGGGUUCGAAGGCAGCUCAGCGGCUACGAUACACUGGCU-GUCGAGCGGUUGGGUUGAUGUUCGAACAAAUUGU------------ ------------((((....(((((......)).)))(((((((.((((((((((.(((.(((((.......)-))))))).))))))))).).)))))))...))))------------ ( -38.20, z-score = -2.94, R) >droEre2.scaffold_4929 24406137 95 + 26641161 ------------ACGGCUUGCCGACGGCCAAGUUCGAGUUCGAGUACAGCUCAAUAGCGACGAUCCACUGGCGUGUCUAGCUGUUGGGCUGAUGU-CGAACAAGUUGC------------ ------------.(((((((.(((((......((((....))))..((((((((((((((((..((...))..))))..)))))))))))).)))-))..))))))).------------ ( -37.50, z-score = -2.08, R) >droYak2.chrX_random 1450020 96 - 1802292 ------------ACAGAUCGCUGACGGUUUAAUUCAAAUUCGAAGGCAACUCAACAGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAUGUUCGAACAAGUUGC------------ ------------..((((((....))))))........((((((.((((((((((.((...(((((......)))))..)).))))))))).).))))))........------------ ( -30.50, z-score = -2.04, R) >droSec1.super_65 166774 120 + 171001 UAUAACCAAAUUACGGAUUGCCGACGGUCGAGUACAAUUUCGAAAGCAGCUCAACGGCUACGAUACACUGGCGUGUCGAGCGGUUGGGUUGAUGUUCGAACAGAUUGCCGACGAUCGAGU .............(((....))).((((((....((((((((((..(((((((((.(((.((((((......))))))))).)))))))))...)))))...)))))....))))))... ( -45.90, z-score = -3.57, R) >droSim1.chr2h_random 560874 106 - 3178526 UAUAACCAAAUUACGGAUUGCCGACGGUUGAGUGUGGGUUCGAGAGCAGCUCAACGGCUACGACACACUGGC--GUCGAGCGGUUGUGUUGAUGUUCGAACAAAUUGU------------ .....(((.....(((....)))((......)).)))(((((((..((((.((((.(((.((((.(....).--))))))).)))).))))...))))))).......------------ ( -36.90, z-score = -2.04, R) >consensus ____________ACGGAUUGCCGACGGUCGAGUACGAGUUCGAAAGCAGCUCAACGGCUACGAUACACUGGCGUGUCGAGCGGUUGGGUUGAUGUUCGAACAAAUUGC____________ .............(((....)))..............(((((((..(((((((((.(((.((((..........))))))).)))))))))...)))))))................... (-25.76 = -26.28 + 0.52)


| Location | 1,848,829 – 1,848,927 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.82 |
| Shannon entropy | 0.41939 |
| G+C content | 0.52282 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -25.76 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 1848829 98 + 2555491 ---------ACAGAUUGCCGACGGUCGAGUACGGGUUCGAAGGCAGCUCAGCGGCUACGAUACACUGGCU-GUCGAGCGGUUGGGUUGAUGUUCGAACAAAUUGUGCU------------ ---------...(((((....))))).((((((.(((((((.((((((((((.(((.(((((.......)-))))))).))))))))).).)))))))....))))))------------ ( -40.60, z-score = -3.05, R) >droEre2.scaffold_4929 24406137 98 + 26641161 ---------ACGGCUUGCCGACGGCCAAGUUCGAGUUCGAGUACAGCUCAAUAGCGACGAUCCACUGGCGUGUCUAGCUGUUGGGCUGAUGU-CGAACAAGUUGCACU------------ ---------.(((((((.(((((......((((....))))..((((((((((((((((..((...))..))))..)))))))))))).)))-))..)))))))....------------ ( -37.50, z-score = -1.77, R) >droYak2.chrX_random 1450020 99 - 1802292 ---------ACAGAUCGCUGACGGUUUAAUUCAAAUUCGAAGGCAACUCAACAGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAUGUUCGAACAAGUUGCGCU------------ ---------......(((.(((((......))...((((((.((((((((((.((...(((((......)))))..)).))))))))).).))))))...))))))..------------ ( -31.20, z-score = -1.62, R) >droSec1.super_65 166777 120 + 171001 AACCAAAUUACGGAUUGCCGACGGUCGAGUACAAUUUCGAAAGCAGCUCAACGGCUACGAUACACUGGCGUGUCGAGCGGUUGGGUUGAUGUUCGAACAGAUUGCCGACGAUCGAGUACA ..........(((....))).((((((....((((((((((..(((((((((.(((.((((((......))))))))).)))))))))...)))))...)))))....))))))...... ( -45.90, z-score = -3.33, R) >droSim1.chr2h_random 560877 106 - 3178526 AACCAAAUUACGGAUUGCCGACGGUUGAGUGUGGGUUCGAGAGCAGCUCAACGGCUACGACACACUGGC--GUCGAGCGGUUGUGUUGAUGUUCGAACAAAUUGUGCU------------ ..(((.....(((....)))((......)).)))(((((((..((((.((((.(((.((((.(....).--))))))).)))).))))...)))))))..........------------ ( -36.90, z-score = -1.37, R) >consensus _________ACGGAUUGCCGACGGUCGAGUACGAGUUCGAAAGCAGCUCAACGGCUACGAUACACUGGCGUGUCGAGCGGUUGGGUUGAUGUUCGAACAAAUUGCGCU____________ ..........(((....)))..............(((((((..(((((((((.(((.((((..........))))))).)))))))))...)))))))...................... (-25.76 = -26.28 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:37 2011