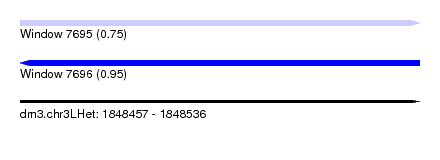
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,848,457 – 1,848,536 |
| Length | 79 |
| Max. P | 0.945505 |

| Location | 1,848,457 – 1,848,536 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 52.11 |
| Shannon entropy | 0.63298 |
| G+C content | 0.35095 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -3.47 |
| Energy contribution | -4.25 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
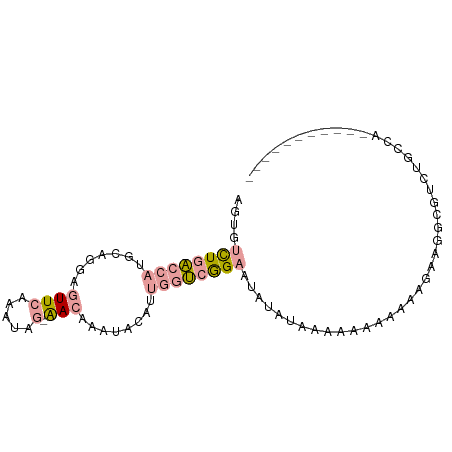
>dm3.chr3LHet 1848457 79 + 2555491 AGUGUCUGACCAUGCAGGAGUUCAAAUAG-AACAAAUACAUUGGUCGGAAUAUAUAAAAAAACAAAGAAGACGUCUGCCA----------- ....((((((((((.....((((.....)-))).....)).))))))))...............................----------- ( -14.70, z-score = -1.21, R) >droSec1.super_65 166473 79 + 171001 AGUUUCUGACCAUGUAGGAGUUCAAAGAG-AACAAAUACAUUGGUCAGAAUAUAUCAAAAACAAAAGAAGGCGUCUGCCA----------- ...(((((((((((((...((((.....)-)))...)))).)))))))))...................(((....))).----------- ( -24.60, z-score = -4.90, R) >droVir3.scaffold_12728 471086 91 - 808307 AAUGUUUUUUCAUACAAAAAUUUAAUUUUCGACCGAUCGAUCCUAUGGCAGCUAUAUGAUAUAGCUGUCCGAUAUUAAUAGGAUUUUGCAC ..............(((((.........((((....))))((((((((((((((((...))))))))))........)))))))))))... ( -20.30, z-score = -2.51, R) >consensus AGUGUCUGACCAUGCAGGAGUUCAAAUAG_AACAAAUACAUUGGUCGGAAUAUAUAAAAAAAAAAAGAAGGCGUCUGCCA___________ ....((((((((.............................)))))))).......................................... ( -3.47 = -4.25 + 0.78)

| Location | 1,848,457 – 1,848,536 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 52.11 |
| Shannon entropy | 0.63298 |
| G+C content | 0.35095 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -2.56 |
| Energy contribution | -3.57 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.12 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
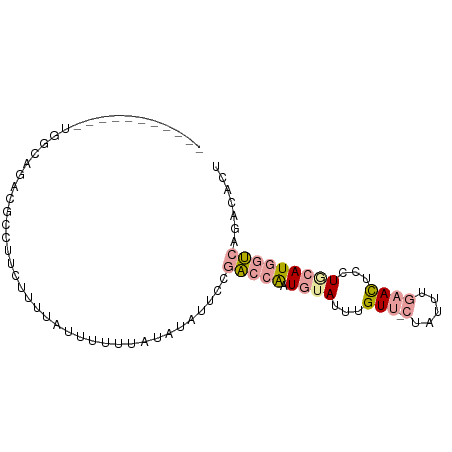
>dm3.chr3LHet 1848457 79 - 2555491 -----------UGGCAGACGUCUUCUUUGUUUUUUUAUAUAUUCCGACCAAUGUAUUUGUU-CUAUUUGAACUCCUGCAUGGUCAGACACU -----------........((((....(((......)))......(((((.((((...(((-(.....))))...)))))))))))))... ( -16.90, z-score = -2.10, R) >droSec1.super_65 166473 79 - 171001 -----------UGGCAGACGCCUUCUUUUGUUUUUGAUAUAUUCUGACCAAUGUAUUUGUU-CUCUUUGAACUCCUACAUGGUCAGAAACU -----------.(((....)))...................(((((((((.((((...(((-(.....))))...)))))))))))))... ( -24.70, z-score = -5.22, R) >droVir3.scaffold_12728 471086 91 + 808307 GUGCAAAAUCCUAUUAAUAUCGGACAGCUAUAUCAUAUAGCUGCCAUAGGAUCGAUCGGUCGAAAAUUAAAUUUUUGUAUGAAAAAACAUU ((((((((((((((.......((.((((((((...))))))))))))))))((((....)))).........))))))))........... ( -24.01, z-score = -3.96, R) >consensus ___________UGGCAGACGCCUUCUUUUAUUUUUUAUAUAUUCCGACCAAUGUAUUUGUU_CUAUUUGAACUCCUGCAUGGUCAGACACU .............................................(((((.((((...(((........)))...)))))))))....... ( -2.56 = -3.57 + 1.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:35 2011