| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,805,291 – 1,805,388 |
| Length | 97 |
| Max. P | 0.978230 |

| Location | 1,805,291 – 1,805,388 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Shannon entropy | 0.42856 |
| G+C content | 0.40685 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -17.10 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
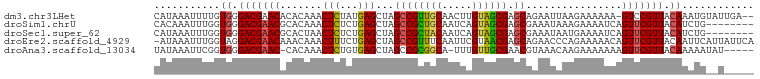
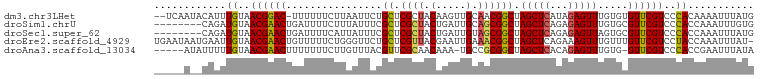
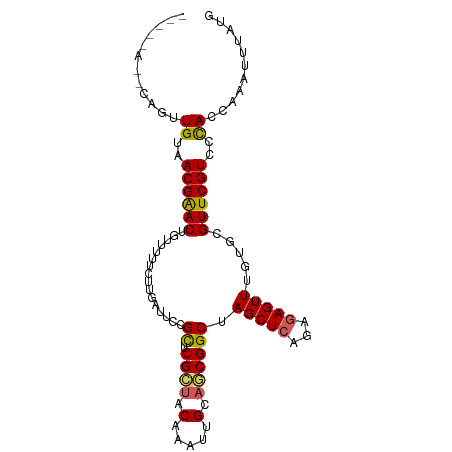
>dm3.chr3LHet 1805291 97 + 2555491 CAUAAAUUUUGUGGGACGAACACACAAACUCUAUGAGCUAGCCGUUGCAACUUGUAGCGAGCAGAAUUAAGAAAAAA-GUCCGUUACAAAUGUAUUGA-- ((.....((((((((((...........(((...)))...((((((((.....)))))).))...............-))))..)))))).....)).-- ( -19.40, z-score = -0.62, R) >droSim1.chrU 12030294 92 + 15797150 CACAAAUUUGGUGGGACGAACGCACAAACUCUCUGAGCUAGCCGCUGCAAUCAGUAGCGAGCGAAAUAAAGAAAAUCAGUUCGUUACAUCUG-------- .........((((..((((((((.((.......)).))..((((((((.....)))))).))................))))))..))))..-------- ( -27.80, z-score = -2.84, R) >droSec1.super_62 172444 92 - 174595 CAUAAAUUUGGUGGGACGAACGCACUAACUCUCUGAGCUAGCCGCUACAAUCAGUAGCGAGCGAAAUAAUGAAAAUCAGUUCGUUACAUCUG-------- .........((((..((((((((.(((.(((...))).))).((((((.....)))))).)).......((.....))))))))..))))..-------- ( -27.10, z-score = -3.08, R) >droEre2.scaffold_4929 23417779 99 + 26641161 -AUAAAUUUGGUAGGACGAACAAACAAACUUUCUGAGCUAGCCGUUUCAAUUCGUAACGAGCAGAACCCAGAAAAACAGUUCGUUACAAUUCAUUAUUCA -......(((....(((((((........((((((..((.((((((.(.....).)))).))))....))))))....))))))).)))........... ( -17.60, z-score = -1.25, R) >droAna3.scaffold_13034 1027812 93 - 2223656 UAUAAAUUCGGUGGGACGAAC-CACAAACUCUGUGAGCUAGCCGCGGCA-UUUGUUGCGAACGUAAACAAGAAAAAAAGUUCGUUACAAAAAUAU----- ..........((((......)-))).....(((((.(....))))))..-(((((.((((((................)))))).))))).....----- ( -21.09, z-score = -1.35, R) >consensus CAUAAAUUUGGUGGGACGAACACACAAACUCUCUGAGCUAGCCGCUGCAAUUAGUAGCGAGCGAAAUAAAGAAAAACAGUUCGUUACAAAUG__U_____ ...........((.(((((((.......(((...)))...((((((((.....)))))).))................))))))).))............ (-17.10 = -16.90 + -0.20)
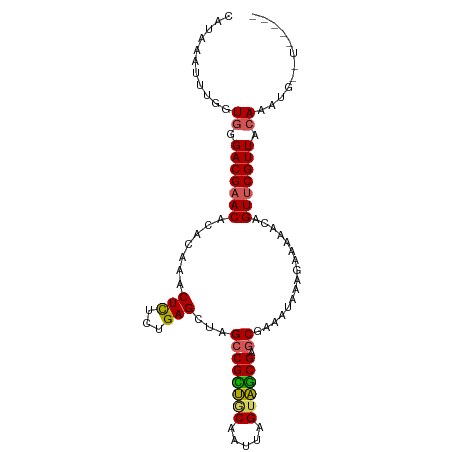
| Location | 1,805,291 – 1,805,388 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.02 |
| Shannon entropy | 0.42856 |
| G+C content | 0.40685 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -13.92 |
| Energy contribution | -13.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 1805291 97 - 2555491 --UCAAUACAUUUGUAACGGAC-UUUUUUCUUAAUUCUGCUCGCUACAAGUUGCAACGGCUAGCUCAUAGAGUUUGUGUGUUCGUCCCACAAAAUUUAUG --........(((((...((((-.........(((((((...((((...((....))...))))...))))))).........)))).)))))....... ( -15.87, z-score = 0.09, R) >droSim1.chrU 12030294 92 - 15797150 --------CAGAUGUAACGAACUGAUUUUCUUUAUUUCGCUCGCUACUGAUUGCAGCGGCUAGCUCAGAGAGUUUGUGCGUUCGUCCCACCAAAUUUGUG --------..((((.((((.((.((((((((.......(((.((..(((....)))..)).)))..)))))))).)).)))))))).............. ( -23.70, z-score = -1.03, R) >droSec1.super_62 172444 92 + 174595 --------CAGAUGUAACGAACUGAUUUUCAUUAUUUCGCUCGCUACUGAUUGUAGCGGCUAGCUCAGAGAGUUAGUGCGUUCGUCCCACCAAAUUUAUG --------..((((.((((.((((((((((........((.((((((.....)))))))).......)))))))))).)))))))).............. ( -29.36, z-score = -3.56, R) >droEre2.scaffold_4929 23417779 99 - 26641161 UGAAUAAUGAAUUGUAACGAACUGUUUUUCUGGGUUCUGCUCGUUACGAAUUGAAACGGCUAGCUCAGAAAGUUUGUUUGUUCGUCCUACCAAAUUUAU- ......((((((...((((((((....(((((((((..((.((((.(.....).)))))).))))))))))))))))).))))))..............- ( -26.00, z-score = -2.78, R) >droAna3.scaffold_13034 1027812 93 + 2223656 -----AUAUUUUUGUAACGAACUUUUUUUCUUGUUUACGUUCGCAACAAA-UGCCGCGGCUAGCUCACAGAGUUUGUG-GUUCGUCCCACCGAAUUUAUA -----.......((..(((((((..............(((..(((.....-))).)))((.(((((...))))).)))-))))))..))........... ( -17.40, z-score = -0.33, R) >consensus _____A__CAGUUGUAACGAACUGUUUUUCUUGAUUCCGCUCGCUACAAAUUGCAGCGGCUAGCUCAGAGAGUUUGUGCGUUCGUCCCACCAAAUUUAUG ............((..((((((................((.((((.(.....).)))))).(((((...))))).....))))))..))........... (-13.92 = -13.80 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:31 2011