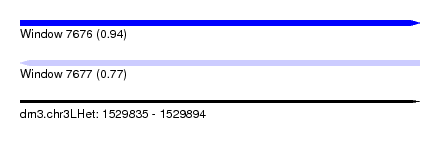
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,529,835 – 1,529,894 |
| Length | 59 |
| Max. P | 0.938232 |

| Location | 1,529,835 – 1,529,894 |
|---|---|
| Length | 59 |
| Sequences | 4 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.35240 |
| G+C content | 0.43220 |
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -11.89 |
| Energy contribution | -10.58 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
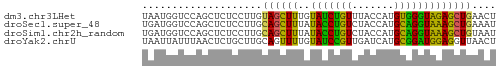
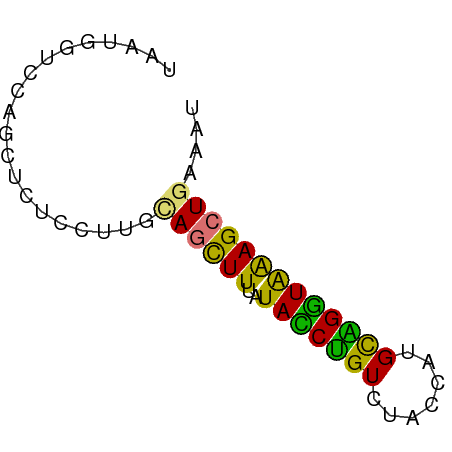
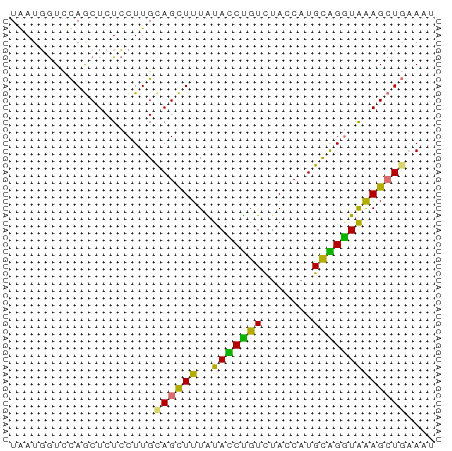
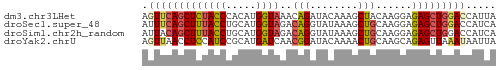
>dm3.chr3LHet 1529835 59 + 2555491 UAAUGGUCCAGCUCUCCUUGUAGCUUUGUAUCUGUUUACCAUGUGGGUAGAGCUGAACU ....(((.(((((((.((..((.....(((......)))..))..)).))))))).))) ( -17.50, z-score = -1.97, R) >droSec1.super_48 78017 59 - 230265 UGAUGGUCCAGCUCUCCUUGCAGCUUUAUACCUGUCUACCAUGCAGGUAAAGCUGAAAU .((.((.....)).))....((((((..(((((((.......))))))))))))).... ( -15.90, z-score = -1.56, R) >droSim1.chr2h_random 741813 59 + 3178526 UGAUGGUCCAGCUCUCCUUGCAGCUUUAUACCUGUCUACCAUGCAGGUAAAGCUGUAAU .((.((.....)).)).(((((((((..(((((((.......)))))))))))))))). ( -19.20, z-score = -2.80, R) >droYak2.chrU 26578999 59 - 28119190 UAAUUAUUUAACUCUGCUUGCAGUUUUGUAUCCGUUGAUCAUGCGGAUGGAGGUUAACU .......(((((((((....)))...(.(((((((.......))))))).))))))).. ( -13.10, z-score = -1.42, R) >consensus UAAUGGUCCAGCUCUCCUUGCAGCUUUAUACCUGUCUACCAUGCAGGUAAAGCUGAAAU ....................((((((..(((((((.......))))))))))))).... (-11.89 = -10.58 + -1.31)
| Location | 1,529,835 – 1,529,894 |
|---|---|
| Length | 59 |
| Sequences | 4 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.35240 |
| G+C content | 0.43220 |
| Mean single sequence MFE | -14.96 |
| Consensus MFE | -9.25 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
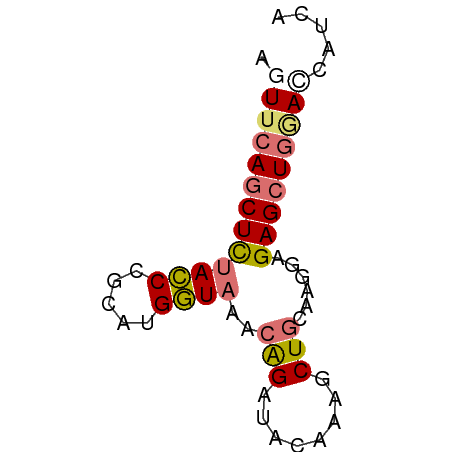
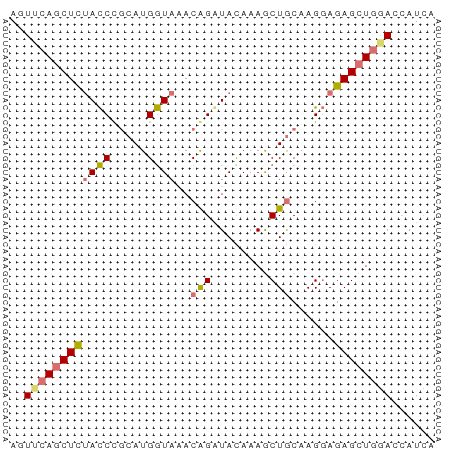
>dm3.chr3LHet 1529835 59 - 2555491 AGUUCAGCUCUACCCACAUGGUAAACAGAUACAAAGCUACAAGGAGAGCUGGACCAUUA .((((((((((.((....((((.............))))...))))))))))))..... ( -18.02, z-score = -3.39, R) >droSec1.super_48 78017 59 + 230265 AUUUCAGCUUUACCUGCAUGGUAGACAGGUAUAAAGCUGCAAGGAGAGCUGGACCAUCA ..(((((((((.(((((.((.....))(((.....))))).))))))))))))...... ( -16.90, z-score = -1.31, R) >droSim1.chr2h_random 741813 59 - 3178526 AUUACAGCUUUACCUGCAUGGUAGACAGGUAUAAAGCUGCAAGGAGAGCUGGACCAUCA ....(((((((.(((((.((.....))(((.....))))).))))))))))........ ( -15.70, z-score = -1.05, R) >droYak2.chrU 26578999 59 + 28119190 AGUUAACCUCCAUCCGCAUGAUCAACGGAUACAAAACUGCAAGCAGAGUUAAAUAAUUA ..(((((....(((((.........)))))......(((....))).)))))....... ( -9.20, z-score = -1.63, R) >consensus AGUUCAGCUCUACCCGCAUGGUAAACAGAUACAAAGCUGCAAGGAGAGCUGGACCAUCA .(((((((((((((.....))))..(((........)))......)))))))))..... ( -9.25 = -10.00 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:20 2011