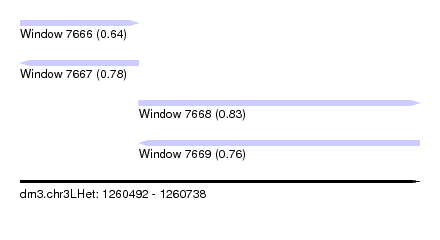
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,260,492 – 1,260,738 |
| Length | 246 |
| Max. P | 0.827686 |

| Location | 1,260,492 – 1,260,565 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 70.31 |
| Shannon entropy | 0.53203 |
| G+C content | 0.53120 |
| Mean single sequence MFE | -22.74 |
| Consensus MFE | -9.49 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638159 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
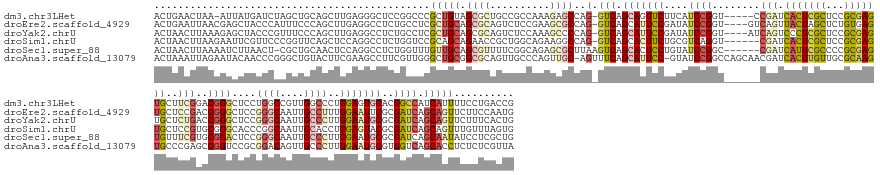
>dm3.chr3LHet 1260492 73 + 2555491 -------UAUAAGUUAACCCAAGCAGUUCUCACGGAGUAAACGUGCAGAAGUGCAAGCCAUA---UCCGGAGGCCUGCACGCG -------...............((((..(((.((((.......((((....)))).......---)))))))..))))..... ( -23.24, z-score = -2.73, R) >droAna3.scaffold_13079 684903 73 - 742402 -------UAUAAGUUAACACGCGCAGGCAGAAUUUAGGAAAAGUGCCAAAUUGCAGACCGGA---UCCGGUGGCCCGCACGCG -------............((.((.(((..(((((.((.......)))))))....((((..---..)))).))).)).)).. ( -17.70, z-score = 0.12, R) >droSec1.super_88 99700 73 - 123530 -------UGCAAGUUAACACAAGCAGUUCUCACGGAGUGAGACUGCCGAAUUGCAAGCUGUA---UCCGGUGGCCUGCACGCG -------.((............(((((.(((((...)))))))))).....((((.((..(.---....)..)).)))).)). ( -23.20, z-score = -0.76, R) >droSim1.chrU 9700766 73 + 15797150 -------UACAAGUUAACACAAGCAGUUCUCACGGAGUGAAACUGCUGAAUUGCAGGCCAUA---UCCGGUGGUCUGCACGCG -------..............(((((((.((((...)))))))))))....((((((((((.---....)))))))))).... ( -27.10, z-score = -3.27, R) >droYak2.chrU 18645223 78 - 28119190 AAAAGGGGAGAAUUUAGCACAUGCGGUUCUCACAGAAAGAAACCGCCGAAUUGCAGGCCAC-----CACGCGCACUGCACGCG ................((....((((((.((.......)))))))).....((((((((..-----...).)).))))).)). ( -19.00, z-score = 0.04, R) >droEre2.scaffold_4929 840794 76 - 26641161 -------UACAAGUUAGCACAGGCGUUUCUUACGGAGAGAAACCGUCGAAUUGCAGGCCACCAUAUCCGGUGGCUUACACGCG -------.........((...((((((((((.....)))))).))))....((.((((((((......)))))))).)).)). ( -26.20, z-score = -3.20, R) >consensus _______UACAAGUUAACACAAGCAGUUCUCACGGAGUGAAACUGCCGAAUUGCAGGCCAUA___UCCGGUGGCCUGCACGCG ......................((..((((...))))..............((((((((((........)))))))))).)). ( -9.49 = -10.13 + 0.64)

| Location | 1,260,492 – 1,260,565 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 70.31 |
| Shannon entropy | 0.53203 |
| G+C content | 0.53120 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -10.69 |
| Energy contribution | -10.38 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777041 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 1260492 73 - 2555491 CGCGUGCAGGCCUCCGGA---UAUGGCUUGCACUUCUGCACGUUUACUCCGUGAGAACUGCUUGGGUUAACUUAUA------- (.((.((((..(((((((---...(((.((((....)))).)))...)))).)))..)))).)).)..........------- ( -24.50, z-score = -2.11, R) >droAna3.scaffold_13079 684903 73 + 742402 CGCGUGCGGGCCACCGGA---UCCGGUCUGCAAUUUGGCACUUUUCCUAAAUUCUGCCUGCGCGUGUUAACUUAUA------- (((((((((((.((((..---..))))....(((((((........)))))))..)))))))))))..........------- ( -27.60, z-score = -3.17, R) >droSec1.super_88 99700 73 + 123530 CGCGUGCAGGCCACCGGA---UACAGCUUGCAAUUCGGCAGUCUCACUCCGUGAGAACUGCUUGUGUUAACUUGCA------- .((((((((((.......---....)))))))....(((((((((((...))))).))))))..........))).------- ( -24.60, z-score = -1.20, R) >droSim1.chrU 9700766 73 - 15797150 CGCGUGCAGACCACCGGA---UAUGGCCUGCAAUUCAGCAGUUUCACUCCGUGAGAACUGCUUGUGUUAACUUGUA------- (((((((((.(((.....---..))).)))))....(((((((((((...))))).))))))))))..........------- ( -21.90, z-score = -1.33, R) >droYak2.chrU 18645223 78 + 28119190 CGCGUGCAGUGCGCGUG-----GUGGCCUGCAAUUCGGCGGUUUCUUUCUGUGAGAACCGCAUGUGCUAAAUUCUCCCCUUUU (((((((...)))))))-----.......(((.....((((((.(((.....)))))))))...)))................ ( -23.50, z-score = -0.44, R) >droEre2.scaffold_4929 840794 76 + 26641161 CGCGUGUAAGCCACCGGAUAUGGUGGCCUGCAAUUCGACGGUUUCUCUCCGUAAGAAACGCCUGUGCUAACUUGUA------- .(((((((.(((((((....))))))).)))..(((.((((.......))))..)))))))...............------- ( -25.00, z-score = -1.97, R) >consensus CGCGUGCAGGCCACCGGA___UAUGGCCUGCAAUUCGGCAGUUUCACUCCGUGAGAACUGCUUGUGUUAACUUGUA_______ .(((((((((((............)))))))).................(....)...)))...................... (-10.69 = -10.38 + -0.30)
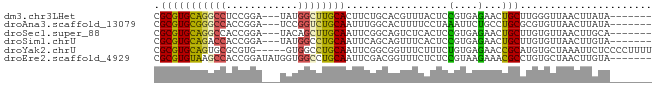
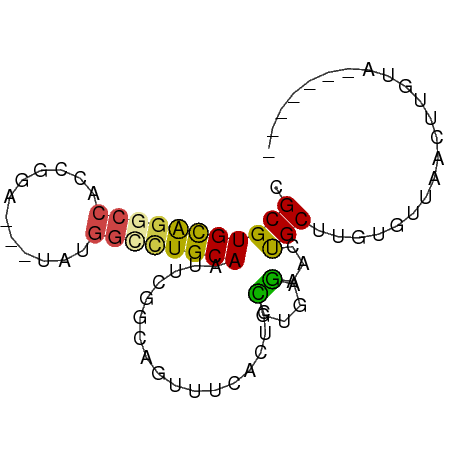
| Location | 1,260,565 – 1,260,738 |
|---|---|
| Length | 173 |
| Sequences | 6 |
| Columns | 180 |
| Reading direction | forward |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.64663 |
| G+C content | 0.57503 |
| Mean single sequence MFE | -70.17 |
| Consensus MFE | -23.05 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 1260565 173 + 2555491 CGGUCAGGAAAAUGAUGGCCGUGCACUCCAGGGCCAACGGCCAGGAGCCCGUCCGAAGCACUCGCGGAGCGAGUGAUCGG-----ACCGGAUGAAGAACUGCUGAC-CUGGCUCUUUGGCGGCAGCGCUACAGCGGGCCGGAGCCCUCAAGCUGCAGCUAGAUCAUAAU-UUAGUUCAGU .((((((........(((((((...((....))...)))))))....((.((((((..(((((((...))))))).))))-----)).))...........)))))-).(((((.....((((..(((....))).))))))))).....((((.((((((((....))-)))))))))) ( -75.20, z-score = -1.94, R) >droEre2.scaffold_4929 840870 175 - 26641161 CAUUGGAAGAACUGCUGAUCGCGAAUUCCAAAGGCAAUUGCCCGGAGCCCGGUCGGAGCACUCACAGAGCUAGUAACUGAC----ACCGGAUAUCGGAAUGCUGAC-CUGGCGCUUCGGAGACUGCGCUGCAGCGAGGCAGAGGCCUCAAGCUGGGAAAUGGGUAGCUCGUUAAUUCAGU .((((((..(((.((((..((((..((((...(((....))).((((((((((((((((.(.....).)))..........----.(((.....)))....)))))-).)).)))))))))..))))...))))(((((....))))).(((((.........))))).)))..)))))) ( -60.80, z-score = -0.03, R) >droYak2.chrU 18645301 175 - 28119190 CAGUGAAAGAACUGCUGAUCGCGCAUUCCAAGGGCAAUUGCCCGGAGCCCGGUCAGAGCACUCGCGGAGCGAGGGACUGAU----ACCGGAUAUCGGAAUGCUGAC-CUGGGGCUUUGGAGACUGCGCUGCAGCGAGGCAGAGGCCUCAAGCUGGGAAACGGGUAGCUCUUUAAGUUAGU .....(((((.((((((...(((((((((..((((....))))(((((((((((((....(((((...)))))...(((((----(.....))))))....)))))-)..)))))))))))..)))))..))))(((((....)))))..(((.(....).))))).)))))........ ( -76.50, z-score = -2.78, R) >droSim1.chrU 9700839 173 + 15797150 CACUAAACAAACUGCUGAUCGCGUACUCCAGGUGCAAUUGCCGGGUGCCCGCACGGAGCACUCGCGGAGCGAGUGAUCG------ACCUGACGCAGAAGUGCUGAC-CUGGCCUUCUGCCAGCGGUUCUGCUGCGGACCAGAGGCCUGGAGCUGAACCGGGAACGAAUUCUUAAGUUAGU ...............(((((((...((((.(((......)))((((((((....)).))))))..))))...)))))))------..((((((((((((.(((...-..))))))))))...((((((.(((.(((.((...)).))).))).))))))...............))))). ( -69.90, z-score = -1.77, R) >droSec1.super_88 99773 173 - 123530 CAGCGAGGAUAUUGCUGAUCGCGCAUUCCAAGGGCAAUUGCCCGGAGUCCGCACGAAACACUCGCGGGGCGAAUGAUCG------GCCGGAUACAGGAGUGCUGACUUAAGCGCUCUGCCGAAAACGCUGCAACAAACCAGAGGCCUGGAGUUGCAGCG-AGUUAAGAUUUUAAGUUAGU ((((((.....)))))).((((..(((((..((((....)))))))))((((...........)))).))))....(((------((.(....).((((((((......)))))))))))))...(((((((((...((((....)))).)))))))))-.................... ( -69.70, z-score = -3.17, R) >droAna3.scaffold_13079 684976 178 - 742402 UAACGAGAGAGGUGCUGACCACGCAUUCCAAGGGCAACUGUCCGCGGACCGGCUCGGGCACUUGCGCAACAAGUGAUCGUUGCUGGCCGGAUAC-GGAAUGCUGAAACU-GCAACUGGGCAACUGCGCCGCAGCCCAACGAAGGCUUCGAAGUACAGCCCGGGUUGUAUUCUAAUUUAGU .....((((.(((....)))..(((((((..((((....)))).(((.(((((.((..((((((.....))))))..))..)))))))).....-)))))))......(-((((((((((...(((..((.((((.......)))).))..)))..)))).)))))))))))........ ( -68.90, z-score = -1.56, R) >consensus CAGUGAAAAAACUGCUGAUCGCGCAUUCCAAGGGCAAUUGCCCGGAGCCCGGACGGAGCACUCGCGGAGCGAGUGAUCG______ACCGGAUACAGGAAUGCUGAC_CUGGCGCUUUGGCGACUGCGCUGCAGCGAACCAGAGGCCUCAAGCUGCAGCACGGGUAGAAUUUUAAGUUAGU .............(((((....(((((((....((....))((((........(((..(((((((...))))))).))).......)))).....))))))).......(((.(((((....((((...)))).....)))))))).............................))))) (-23.05 = -24.73 + 1.68)
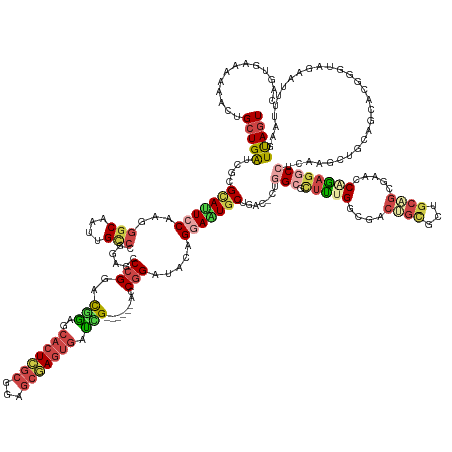
| Location | 1,260,565 – 1,260,738 |
|---|---|
| Length | 173 |
| Sequences | 6 |
| Columns | 180 |
| Reading direction | reverse |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.64663 |
| G+C content | 0.57503 |
| Mean single sequence MFE | -64.93 |
| Consensus MFE | -21.54 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.51 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 1260565 173 - 2555491 ACUGAACUAA-AUUAUGAUCUAGCUGCAGCUUGAGGGCUCCGGCCCGCUGUAGCGCUGCCGCCAAAGAGCCAG-GUCAGCAGUUCUUCAUCCGGU-----CCGAUCACUCGCUCCGCGAGUGCUUCGGACGGGCUCCUGGCCGUUGGCCCUGGAGUGCACGGCCAUCAUUUUCCUGACCG ...(((((..-...........((((((((....((((....))))))))))))((((((((......))..)-).))))))))).....(((((-----((((.(((((((...)))))))..))))))(.(((((.((((...))))..))))).).))).................. ( -75.40, z-score = -2.68, R) >droEre2.scaffold_4929 840870 175 + 26641161 ACUGAAUUAACGAGCUACCCAUUUCCCAGCUUGAGGCCUCUGCCUCGCUGCAGCGCAGUCUCCGAAGCGCCAG-GUCAGCAUUCCGAUAUCCGGU----GUCAGUUACUAGCUCUGUGAGUGCUCCGACCGGGCUCCGGGCAAUUGCCUUUGGAAUUCGCGAUCAGCAGUUCUUCCAAUG ...(((((...(((((...........)))))(((((....)))))((((..(((.(((.((((((..(((.(-((((((((((((((((...))----)))(((.....)))..).)))))))..)))).)))....(((....)))))))))))))))...)))))))))........ ( -56.80, z-score = -0.32, R) >droYak2.chrU 18645301 175 + 28119190 ACUAACUUAAAGAGCUACCCGUUUCCCAGCUUGAGGCCUCUGCCUCGCUGCAGCGCAGUCUCCAAAGCCCCAG-GUCAGCAUUCCGAUAUCCGGU----AUCAGUCCCUCGCUCCGCGAGUGCUCUGACCGGGCUCCGGGCAAUUGCCCUUGGAAUGCGCGAUCAGCAGUUCUUUCACUG ........((((((((................(((((....)))))((((..(((((...((((((((((..(-(((((....(((.....))).----...(((..(((((...))))).))))))))))))))..((((....))))))))).)))))...))))))))))))..... ( -69.80, z-score = -4.07, R) >droSim1.chrU 9700839 173 - 15797150 ACUAACUUAAGAAUUCGUUCCCGGUUCAGCUCCAGGCCUCUGGUCCGCAGCAGAACCGCUGGCAGAAGGCCAG-GUCAGCACUUCUGCGUCAGGU------CGAUCACUCGCUCCGCGAGUGCUCCGUGCGGGCACCCGGCAAUUGCACCUGGAGUACGCGAUCAGCAGUUUGUUUAGUG (((((.....((((.((....)))))).((((((((......(((((((((((((..((((((..........-))))))..)))))).......------.((.(((((((...))))))).))..))))))).....((....)).))))))))..((.....)).......))))). ( -63.20, z-score = -0.54, R) >droSec1.super_88 99773 173 + 123530 ACUAACUUAAAAUCUUAACU-CGCUGCAACUCCAGGCCUCUGGUUUGUUGCAGCGUUUUCGGCAGAGCGCUUAAGUCAGCACUCCUGUAUCCGGC------CGAUCAUUCGCCCCGCGAGUGUUUCGUGCGGACUCCGGGCAAUUGCCCUUGGAAUGCGCGAUCAGCAAUAUCCUCGCUG ....................-(((((((((.((((....))))...)))))))))...(((((.(((.(((......))).))).........))------))).(((((((...)))))))..(((((((...(((((((....))))..))).))))))).((((.........)))) ( -63.60, z-score = -2.91, R) >droAna3.scaffold_13079 684976 178 + 742402 ACUAAAUUAGAAUACAACCCGGGCUGUACUUCGAAGCCUUCGUUGGGCUGCGGCGCAGUUGCCCAGUUGC-AGUUUCAGCAUUCC-GUAUCCGGCCAGCAACGAUCACUUGUUGCGCAAGUGCCCGAGCCGGUCCGCGGACAGUUGCCCUUGGAAUGCGUGGUCAGCACCUCUCUCGUUA ....................((((((.(((.((((((..(..((((((.((......)).))))))..).-.))))).(((((((-((..((((((.(((((((....)))))))((....))..).)))))...))((........))..)))))))).))))))).)).......... ( -60.80, z-score = -0.11, R) >consensus ACUAAAUUAAAAAGCUACCCGCGCUGCAGCUCGAGGCCUCUGGCUCGCUGCAGCGCAGUCGCCAAAGCGCCAG_GUCAGCAUUCCUGUAUCCGGU______CGAUCACUCGCUCCGCGAGUGCUCCGACCGGGCUCCGGGCAAUUGCCCUUGGAAUGCGCGAUCAGCAGUUCUCUCACUG ..............................................(((((.((((..........))))....(((.(((((((.((((((((............((((((...))))))((((.....)))).)))))....)))....)))))))..)))..))))).......... (-21.54 = -22.30 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:14 2011