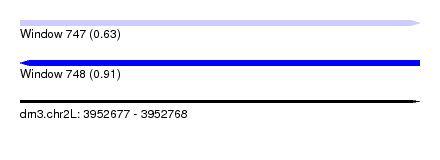
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,952,677 – 3,952,768 |
| Length | 91 |
| Max. P | 0.910813 |

| Location | 3,952,677 – 3,952,768 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.84 |
| Shannon entropy | 0.49354 |
| G+C content | 0.51210 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.34 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
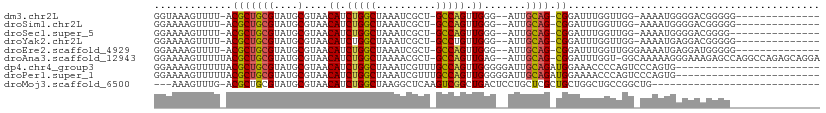
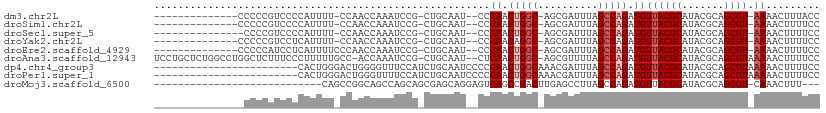
>dm3.chr2L 3952677 91 + 23011544 GGUAAAGUUUU-ACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGCU-GCCAGUUGGG--AUUGCAG-CGGAUUUGGUUGG-AAAAUGGGGACGGGGG-------------- .........((-((((.......))))))..((..(((((((((((-((.(((....--)))))))-).))))))))..)-)...((....))....-------------- ( -30.50, z-score = -2.56, R) >droSim1.chr2L 3907789 91 + 22036055 GGAAAAGUUUU-ACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGCU-GCCAGUUGGG--AUUGCAG-CGGAUUUGGUUGG-AAAAUGGGGACGGGGG-------------- .........((-((((.......))))))..((..(((((((((((-((.(((....--)))))))-).))))))))..)-)...((....))....-------------- ( -30.50, z-score = -2.72, R) >droSec1.super_5 2055452 90 + 5866729 GGAAAAGUUUU-ACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGCU-GCCAGUUGGG--AUUGCAG-CGGAUUUGGUUGG-AAAAUGGGGACGGGG--------------- .........((-((((.......))))))..((..(((((((((((-((.(((....--)))))))-).))))))))..)-)...((....))...--------------- ( -30.50, z-score = -2.81, R) >droYak2.chr2L 3967155 91 + 22324452 GGAAAAGUUUU-ACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGCU-GCCUGUUGGG--AUUGCAG-CGGAUUUGGUUGG-AAAAUGAGGACGGGGG-------------- ......(((((-((((.......))))))..((..(((((((((((-((........--...))))-).))))))))..)-).......))).....-------------- ( -28.50, z-score = -1.96, R) >droEre2.scaffold_4929 4010674 92 + 26641161 GGAAAAGUUUU-ACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGCU-GCCAGUUGGG--AUUGCAG-CGGAUUUGGUUGGGAAAAUGAGGAUGGGGG-------------- .........((-((((.......))))))..((..(((((((((((-((.(((....--)))))))-).))))))))..))................-------------- ( -27.80, z-score = -1.89, R) >droAna3.scaffold_12943 499186 106 - 5039921 GGAAAAGUUUUUACGCUGCGUAUGCGUAACAUCUGGCUAAAACGCU-GCCAGUUGAG--AUUGCAG-CGGAUUUGGU-GGCAAAAAGGGAAAGAGCCAGGCCAGAGCAGGA ..........((((((.......))))))..(((((((....((((-((.(((....--)))))))-)).......(-(((.............)))))))))))...... ( -32.82, z-score = -0.87, R) >dp4.chr4_group3 9630459 87 - 11692001 GGAAAAGUUUUUACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGUUUGCCAGUUGGGGGAUUGCAGAUGGAAACCCCAGUCCCAGUG------------------------ ..........((((((.......))))))((.(((((..........))))).)).(((((((.....(....)..)))))))....------------------------ ( -26.70, z-score = -1.21, R) >droPer1.super_1 6745588 87 - 10282868 GGAAAAGUUUUUACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGUUUGCCAGUUGGGGGAUUGCAGAUGGAAAACCCAGUCCCAGUG------------------------ ..........((((((.......))))))((.(((((..........))))).)).(((((((.....((....)))))))))....------------------------ ( -25.40, z-score = -1.00, R) >droMoj3.scaffold_6500 1966386 79 + 32352404 ---AAAGUUUG-ACGCUGCGUAUGCGUAACAUCUGGCUAAGGCUCAAGUCGGCUGACUCCUGCUCGCUGCUGGCUGCCGGCUG---------------------------- ---..(((..(-(.(.((((....)))).).))..)))........(((((((.....((.((.....)).))..))))))).---------------------------- ( -23.00, z-score = 0.86, R) >consensus GGAAAAGUUUU_ACGCUGCGUAUGCGUAACAUCUGGCUAAAUCGCU_GCCAGUUGGG__AUUGCAG_CGGAUUUGGUUGG_AAAAUGGGGACGGGG_______________ .............(((((((.........((.(((((..........))))).))......))))).)).......................................... (-13.40 = -13.34 + -0.06)
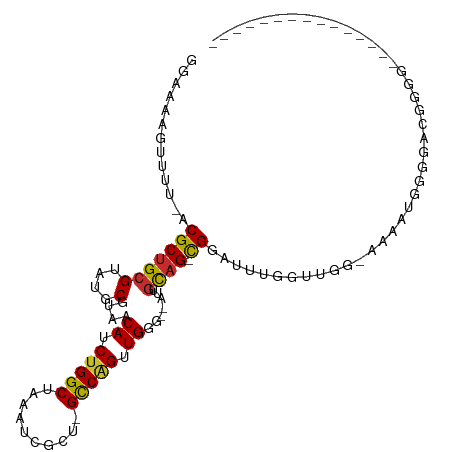
| Location | 3,952,677 – 3,952,768 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.84 |
| Shannon entropy | 0.49354 |
| G+C content | 0.51210 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -9.04 |
| Energy contribution | -9.64 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3952677 91 - 23011544 --------------CCCCCGUCCCCAUUUU-CCAACCAAAUCCG-CUGCAAU--CCCAACUGGC-AGCGAUUUAGCCAGAUGUUACGCAUACGCAGCGU-AAAACUUUACC --------------................-...........((-((((...--.....(((((-.........)))))(((.....)))..)))))).-........... ( -19.70, z-score = -3.28, R) >droSim1.chr2L 3907789 91 - 22036055 --------------CCCCCGUCCCCAUUUU-CCAACCAAAUCCG-CUGCAAU--CCCAACUGGC-AGCGAUUUAGCCAGAUGUUACGCAUACGCAGCGU-AAAACUUUUCC --------------................-...........((-((((...--.....(((((-.........)))))(((.....)))..)))))).-........... ( -19.70, z-score = -3.29, R) >droSec1.super_5 2055452 90 - 5866729 ---------------CCCCGUCCCCAUUUU-CCAACCAAAUCCG-CUGCAAU--CCCAACUGGC-AGCGAUUUAGCCAGAUGUUACGCAUACGCAGCGU-AAAACUUUUCC ---------------...............-...........((-((((...--.....(((((-.........)))))(((.....)))..)))))).-........... ( -19.70, z-score = -3.16, R) >droYak2.chr2L 3967155 91 - 22324452 --------------CCCCCGUCCUCAUUUU-CCAACCAAAUCCG-CUGCAAU--CCCAACAGGC-AGCGAUUUAGCCAGAUGUUACGCAUACGCAGCGU-AAAACUUUUCC --------------................-......(((((.(-((((...--........))-)))))))).........((((((.......))))-))......... ( -16.50, z-score = -2.07, R) >droEre2.scaffold_4929 4010674 92 - 26641161 --------------CCCCCAUCCUCAUUUUCCCAACCAAAUCCG-CUGCAAU--CCCAACUGGC-AGCGAUUUAGCCAGAUGUUACGCAUACGCAGCGU-AAAACUUUUCC --------------............................((-((((...--.....(((((-.........)))))(((.....)))..)))))).-........... ( -19.70, z-score = -3.57, R) >droAna3.scaffold_12943 499186 106 + 5039921 UCCUGCUCUGGCCUGGCUCUUUCCCUUUUUGCC-ACCAAAUCCG-CUGCAAU--CUCAACUGGC-AGCGUUUUAGCCAGAUGUUACGCAUACGCAGCGUAAAAACUUUUCC ......((((((.((((.............)))-)...((..((-((((...--........))-))))..)).))))))..((((((.......)))))).......... ( -30.12, z-score = -2.78, R) >dp4.chr4_group3 9630459 87 + 11692001 ------------------------CACUGGGACUGGGGUUUCCAUCUGCAAUCCCCCAACUGGCAAACGAUUUAGCCAGAUGUUACGCAUACGCAGCGUAAAAACUUUUCC ------------------------....((((.((.((.......)).)).)))).((.(((((..........))))).))((((((.......)))))).......... ( -23.00, z-score = -1.24, R) >droPer1.super_1 6745588 87 + 10282868 ------------------------CACUGGGACUGGGUUUUCCAUCUGCAAUCCCCCAACUGGCAAACGAUUUAGCCAGAUGUUACGCAUACGCAGCGUAAAAACUUUUCC ------------------------...((((...(((((..........))))))))).(((((..........)))))...((((((.......)))))).......... ( -20.40, z-score = -0.85, R) >droMoj3.scaffold_6500 1966386 79 - 32352404 ----------------------------CAGCCGGCAGCCAGCAGCGAGCAGGAGUCAGCCGACUUGAGCCUUAGCCAGAUGUUACGCAUACGCAGCGU-CAAACUUU--- ----------------------------.....(((.((.....))..((..(((((....)))))..))....))).((((((.((....)).)))))-).......--- ( -20.60, z-score = 0.26, R) >consensus _______________CCCCGUCCCCAUUUU_CCAACCAAAUCCG_CUGCAAU__CCCAACUGGC_AGCGAUUUAGCCAGAUGUUACGCAUACGCAGCGU_AAAACUUUUCC .............................................((((..........(((((.((....)).)))))(((.....)))..))))............... ( -9.04 = -9.64 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:04 2011