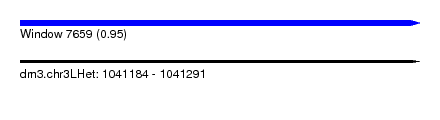
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,041,184 – 1,041,291 |
| Length | 107 |
| Max. P | 0.954634 |

| Location | 1,041,184 – 1,041,291 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 66.34 |
| Shannon entropy | 0.69088 |
| G+C content | 0.51309 |
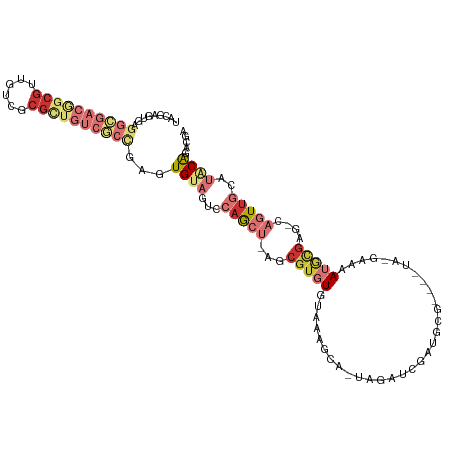
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -15.18 |
| Energy contribution | -18.40 |
| Covariance contribution | 3.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
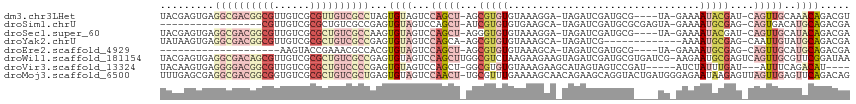
>dm3.chr3LHet 1041184 107 + 2555491 UACGAGUGAGGCGACGGCGUUGUCGCGUUGUCGCCUAGUGUAGUCCAGCU-AGCGUGUGUAAAGGA-UAGAUCGAUGCG----UA-GAAAAUACGAU-CAGUUGCAAACAGACGU ((((....((((((((((((....))))))))))))..))))........-.((((.(((....(.-..(((.(((.((----((-.....))))))-).))).)..))).)))) ( -35.50, z-score = -1.58, R) >droSim1.chrU 9457016 94 - 15797150 -----------------CGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCU-AUCGUGUGUGAAGCA-UAGAUCGAUGCGCGAGUA-GAAAAUGCGAG-CAGUGACAUGCAGACGA -----------------..(((((..((((((((.(..((((......((-(((((((((((....-....)).)))))))).))-)....))))..-).)))))).)).))))) ( -30.10, z-score = -0.17, R) >droSec1.super_60 94623 107 + 163194 UACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCU-AGGGUGUGUAAAGGA-UAGAUCGAUGCG----UA-GAAAAUACGAU-CAGUUGCAUACAGACGA ..((.....(((((((((((....)))))))))))...((((...(((((-..(((.(((.....)-)).)))(((.((----((-.....))))))-))))))..))))..)). ( -38.20, z-score = -2.41, R) >droYak2.chrU 1121591 99 + 28119190 UAUAAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCA-AGCGUGUGUAAAGCA-UAGAUCG-------------AAAAUGCGAG-CAAUUGUAUGCAGACGA ((((.....(((((((((((....)))))))))))...))))(((..(((-.(((..(((...(((-(......-------------...))))..)-))..))).))).))).. ( -35.50, z-score = -2.08, R) >droEre2.scaffold_4929 4582357 87 - 26641161 --------------------AAGUACCGAAACGCCACGUGUAGUCCAGCU-AGCGUGUGUAAAGCA-UAGAUCGAUGCG----UA-GAAAAUGCGAG-CAGUUGCAUGCAGACGA --------------------...........(((..(((((((.(..(((-.((((.......(((-(......)))).----..-....)))).))-).))))))))..).)). ( -19.24, z-score = 0.93, R) >droWil1.scaffold_181154 1830365 114 + 4610121 UACGAGUGAGGCGACAGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCUUGGCGUCUAAGAAGAAGUAGAUCGAUGCGUGAUCG-AAGAAUGCGAGUCAGUUGCGUUCGGAUAA ((((.....(((((((((((....)))))))))))...))))((((.((....((((((........)))).))..)).......-..((((((((.....)))))))))))).. ( -43.20, z-score = -2.16, R) >droVir3.scaffold_13324 1920230 102 - 2960039 UACAAGUGAGGGGACGGCGUUGUCGCGCUGUCCCCGAGUGUAGUCCAGCU-GGCGUGUGUAAAGAAGCAUAGUAGUCCGAU-----AUCUAUUUGAU---AUUUCAGACAU---- ((((.....(((((((((((....)))))))))))...))))(((..(((-(.(.(((((......))))))))))..(((-----(((.....)))---)))...)))..---- ( -35.90, z-score = -3.00, R) >droMoj3.scaffold_6500 31239535 114 + 32352404 UUUGAGCGAGGCGACGGCGGUGUCGCGCUGUCGCUGAGUGUAGUCCAACU-UGCGUUUGAAAAGCAACAGAAGCAGGUACUGAUGGGAGAAUAAGAGUUAGUUGAGUUCAGACAG (((((((..((((((((((......))))))))))........(((((((-(((.((((........)))).)))))).....))))..................)))))))... ( -36.30, z-score = -1.26, R) >consensus UACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCU_AGCGUGUGUAAAGCA_UAGAUCGAUGCG____UA_GAAAAUGCGAG_CAGUUGCAUACAGACGA .........((((((((((......))))))))))...((((...(((((...(((((................................)))))....)))))..))))..... (-15.18 = -18.40 + 3.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:05 2011