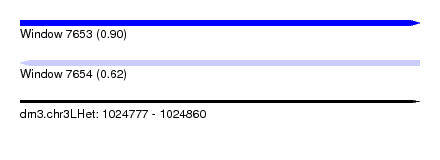
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,024,777 – 1,024,860 |
| Length | 83 |
| Max. P | 0.900334 |

| Location | 1,024,777 – 1,024,860 |
|---|---|
| Length | 83 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 57.83 |
| Shannon entropy | 0.69344 |
| G+C content | 0.44004 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -8.88 |
| Energy contribution | -8.50 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
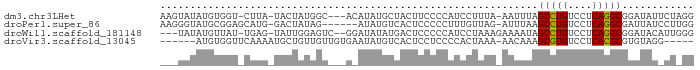
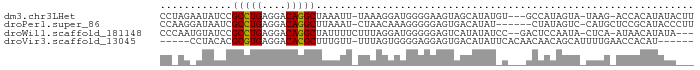
>dm3.chr3LHet 1024777 83 + 2555491 AAGUAUAUGUGGU-CUUA-UACUAUGGC---ACAUAUGCUACUUCCCCAUCCUUUA-AAUUUAGCCUGUCCUCAGGCGGAUAUUCUAGG .((((((((((.(-(...-......)))---)))))))))..........(((...-......(((((....))))).........))) ( -22.07, z-score = -2.17, R) >droPer1.super_86 35294 81 - 225837 AAGGGUAUGCGGAGCAUG-GACUAUAG------AUAUGUCACUCCCCCUUUGUUAG-AUUUAAGCCUGUCCUCAGGCGAUUAUCCUUGG (((((((..(((((....-(((.....------....))).))))...........-......(((((....))))))..))))))).. ( -22.20, z-score = -0.70, R) >droWil1.scaffold_181148 3512982 82 - 5435427 ---UAUAUGUUAU-UGAG-UAUUGGAGUC--GGAUAUAUGACUCCCCCAUCCUAAAGAAAAUAGCCUGUCCUCAGGCGGAUACAUUGGG ---..........-...(-((((((((((--(......)))))))..................(((((....))))).)))))...... ( -22.60, z-score = -1.35, R) >droVir3.scaffold_13045 2045985 77 - 2268007 ------AUGUGGUUCAAAAUGCUGUUGUUGUGAAUAUGUCACUCCUCCCCACUAAA-AACAAAGCGUGUCCUCACGCGUGUAGG----- ------..((((........((....)).((((.....))))......))))....-.(((..(((((....))))).)))...----- ( -14.70, z-score = -0.70, R) >consensus ___UAUAUGUGGU_CAAA_UACUAUAGU___G_AUAUGUCACUCCCCCAUCCUAAA_AAUAAAGCCUGUCCUCAGGCGGAUAUACU_GG ...............................................................(((((....)))))............ ( -8.88 = -8.50 + -0.38)
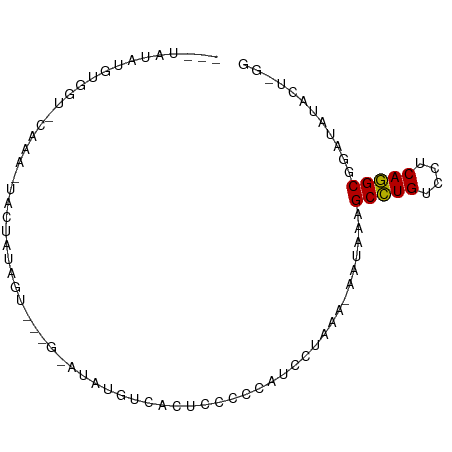
| Location | 1,024,777 – 1,024,860 |
|---|---|
| Length | 83 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 57.83 |
| Shannon entropy | 0.69344 |
| G+C content | 0.44004 |
| Mean single sequence MFE | -19.51 |
| Consensus MFE | -8.07 |
| Energy contribution | -7.70 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 1024777 83 - 2555491 CCUAGAAUAUCCGCCUGAGGACAGGCUAAAUU-UAAAGGAUGGGGAAGUAGCAUAUGU---GCCAUAGUA-UAAG-ACCACAUAUACUU (((....((((((((((....)))))((....-))..))))))))(((((...(((((---(........-....-..))))))))))) ( -16.14, z-score = 0.10, R) >droPer1.super_86 35294 81 + 225837 CCAAGGAUAAUCGCCUGAGGACAGGCUUAAAU-CUAACAAAGGGGGAGUGACAUAU------CUAUAGUC-CAUGCUCCGCAUACCCUU ....((((....(((((....)))))....))-))....(((((((((((((....------.....)))-...))))).....))))) ( -21.20, z-score = -0.91, R) >droWil1.scaffold_181148 3512982 82 + 5435427 CCCAAUGUAUCCGCCUGAGGACAGGCUAUUUUCUUUAGGAUGGGGGAGUCAUAUAUCC--GACUCCAAUA-CUCA-AUAACAUAUA--- ....((((.((((((((((((.((....)).)))))))).))))((((((........--))))))....-....-...))))...--- ( -24.30, z-score = -2.12, R) >droVir3.scaffold_13045 2045985 77 + 2268007 -----CCUACACGCGUGAGGACACGCUUUGUU-UUUAGUGGGGAGGAGUGACAUAUUCACAACAACAGCAUUUUGAACCACAU------ -----.......(((((....)))))...(((-(..((((.(..(..((((.....))))..)..)..))))..)))).....------ ( -16.40, z-score = -0.90, R) >consensus CC_AGAAUAUCCGCCUGAGGACAGGCUAAAUU_UUAAGGAUGGGGGAGUGACAUAU_C___ACUACAGUA_CAUG_ACCACAUAUA___ ............(((((....)))))............................................................... ( -8.07 = -7.70 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:01 2011