| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 1,002,196 – 1,002,351 |
| Length | 155 |
| Max. P | 0.696152 |

| Location | 1,002,196 – 1,002,351 |
|---|---|
| Length | 155 |
| Sequences | 4 |
| Columns | 164 |
| Reading direction | forward |
| Mean pairwise identity | 70.81 |
| Shannon entropy | 0.46869 |
| G+C content | 0.45611 |
| Mean single sequence MFE | -35.39 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.09 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
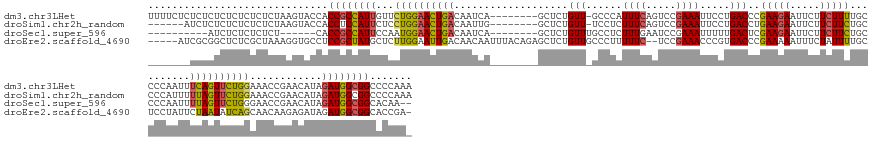
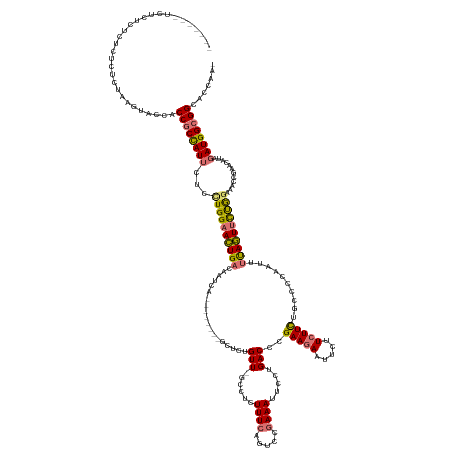
>dm3.chr3LHet 1002196 155 + 2555491 UUUUCUCUCUCUCUCUCUCUCUAAGUACCACCGCCAUUGUUCUGGAACUGACAAUCA--------GCUCUGUU-GCCCAUUUCAGUCCGAAAUUCCUGACCCGAAGAAUUCUUCUUUUGCCCCAAUUUCAGUUCUGGAAACCGAACAUAGAUGGCGGCCCCAAA ..............................((((((((.....(((.((((.(((..--------((......-))..))))))))))((((((........((((....)))).........)))))).((((.(....).))))...))))))))....... ( -35.53, z-score = -2.03, R) >droSim1.chr2h_random 1701456 149 + 3178526 ------AUCUCUCUCUCUCUCUAAGUACCACCUCCAUUCUCCUGGAACUGACAAUUG--------GCUCUGUU-UCCUCUUUCAGUCCGAAAUUCCUGACCUGAAGAAUUCUUCUUCUGCCCCAUUUUUAGUUCUGGAAACCGAACAUAGAUGGCGGCCCCAAA ------......................((..((((......))))..))....(((--------(.......-...(((((..(((.(......).)))..)))))...........(((((((((...((((.(....).))))..)))))).))).)))). ( -30.70, z-score = -1.03, R) >droSec1.super_596 7523 138 - 10499 ----------AUCUCUCUCUCU------CACCGCCAUUCCAAUGGAACUGACAAUCA--------GCUCUGUUUGCCUCUUUGAAUCCGAAAUUUUUGACUCGAAGAAUUCUUCUUCUGCCCCAAUUUUAGUUCUGGGAACCGAACAUAGAUGGCGGCACAA-- ----------............------..((((((((.....(((.(((.....))--------).)))(((((..(((..((((..((((((........(((((.....)))))......)))))).))))..)))..)))))...)))))))).....-- ( -32.24, z-score = -1.47, R) >droEre2.scaffold_4690 18587432 156 + 18748788 -----AUCGCGGCUCUCGCUAAAGGUGCCUCCGCUAUGCUCUUGGAAUUGACAACAAUUUACAGAGCUCUGUUGCCCUUUUUC--UCCGAAACCCGUGACCCGAAAAAUUUCUAUUUUGCUCCUAUUCUAAUAUCAGCAACAAGAGAUAGAUGGCGGCACCGA- -----...(((.....)))....((((((.(((((((.((((((((((((....))))))...((((...((.....((((((--((((.....)).))...)))))).....))...))))..................)))))))))).))).))))))..- ( -43.10, z-score = -2.35, R) >consensus _______UCUCUCUCUCUCUCUAAGUACCACCGCCAUUCUCCUGGAACUGACAAUCA________GCUCUGUU_GCCUCUUUCAGUCCGAAAUUCCUGACCCGAAGAAUUCUUCUUCUGCCCCAAUUUUAGUUCUGGAAACCGAACAUAGAUGGCGGCACCAA_ ..............................((((((((...((((((((((...................(((......((((.....)))).....)))..(((((.....)))))..........))))))))))............))))))))....... (-17.02 = -17.09 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:51:00 2011