
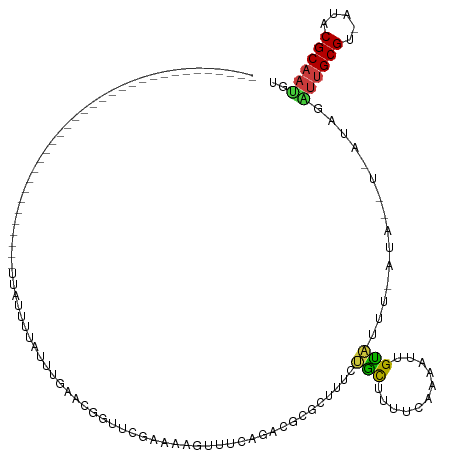
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,945,736 – 3,945,827 |
| Length | 91 |
| Max. P | 0.749635 |

| Location | 3,945,736 – 3,945,827 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 126 |
| Reading direction | forward |
| Mean pairwise identity | 51.29 |
| Shannon entropy | 0.77166 |
| G+C content | 0.37764 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -5.96 |
| Energy contribution | -6.10 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3945736 91 + 23011544 ---------------------------------UUAUUUUAUUUGAACGGUUCGAAAAGUUUCGGACGCGCUUUCUGCUUUUCAGAUUUGUAUUU-AUAUUUCAUAGAUUGCGU-AUACGCAAUGU ---------------------------------.(((...(((((((..(((((((....)))))))..((.....))..)))))))..)))...-...........((((((.-...)))))).. ( -19.90, z-score = -1.46, R) >droSim1.chr2L 3900651 91 + 22036055 ---------------------------------UUAUUUUAUUUGUACGGUUCGAAAAGUUUCUGACGCUCUUUCUGCUUUUCAGAUUUGUAUUU-AUAUUUCAUAGAUUGCGU-AUACGCAAUGU ---------------------------------......(((..((((((.(((((((((....(.....).....))))))).)).))))))..-)))........((((((.-...)))))).. ( -18.50, z-score = -1.97, R) >droSec1.super_5 2048573 80 + 5866729 ---------------------------------UUAUUUUAUUUGACCGGUUCGAAAAGUUUC------------UGCUUUUCAGAUUUGUAUUU-AUAUAUUAUAGAUUGCGUUAUACGCAAUGU ---------------------------------((((..(((...((..((..(((((((...------------.)))))))..))..))....-..)))..))))((((((.....)))))).. ( -16.80, z-score = -2.32, R) >droEre2.scaffold_4929 4003554 92 + 26641161 ---------------------------------UUAUUUUAUUUGAACGGUUCGAAAAGUUUCAGACGCCCUUUCUGCUUUUCCAACUUGUUUUUCAUACUUCAUAGGUUGCGU-AUACGCAACGU ---------------------------------...........((((((((.((((((...((((.......)))))))))).))).)))))..............((((((.-...)))))).. ( -19.80, z-score = -2.22, R) >droAna3.scaffold_12943 492778 117 - 5039921 GCACUUUAUUCAAACAGUUUGAAAAGUUCACCACUUCCACCCAAGGACUUUCGGAUUCAAUCCGCACUCAGUUCCCUUUUUCGAAAAGUGGGAGC--------CACCCUGGCGU-AUACGCAAUGU ..............(((..((....((((.((((((......(((((((..(((((...))))).....)))..)))).......))))))))))--------))..)))(((.-...)))..... ( -24.52, z-score = -0.61, R) >dp4.chr4_group3 9621211 70 - 11692001 ------------------------------------------GCGUAUUAACCGAAGUGUUCUAGAAGUGUUUUCCCUUUU---AAAGCGGGCG----------AACGUUGCGU-AUACGCAAUGU ------------------------------------------..........((...((((.((((((........)))))---).))))..))----------.((((((((.-...)))))))) ( -16.20, z-score = -0.34, R) >droPer1.super_1 6736415 70 - 10282868 ------------------------------------------GCGUAUUAACCGAAGUGUUCUAGAAGUGUUUUCCCUUUU---AAAGCGGGCG----------AACGCUGCGU-AUACGCAAUGU ------------------------------------------((((((....((.(((((((((((((........)))))---)..(....))----------)))))).)).-))))))..... ( -19.20, z-score = -1.14, R) >consensus _________________________________UUAUUUUAUUUGAACGGUUCGAAAAGUUUCAGACGCGCUUUCUGCUUUUCAAAAUUGUAUUU_AUA__U_AUAGAUUGCGU_AUACGCAAUGU ...........................................................................................................((((((.....)))))).. ( -5.96 = -6.10 + 0.14)


| Location | 3,945,736 – 3,945,827 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 51.29 |
| Shannon entropy | 0.77166 |
| G+C content | 0.37764 |
| Mean single sequence MFE | -17.36 |
| Consensus MFE | -4.98 |
| Energy contribution | -5.00 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.531092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3945736 91 - 23011544 ACAUUGCGUAU-ACGCAAUCUAUGAAAUAU-AAAUACAAAUCUGAAAAGCAGAAAGCGCGUCCGAAACUUUUCGAACCGUUCAAAUAAAAUAA--------------------------------- ..((((((...-.))))))...(((.....-.........((((.....))))..(((.((.((((....)))).))))))))..........--------------------------------- ( -15.80, z-score = -1.81, R) >droSim1.chr2L 3900651 91 - 22036055 ACAUUGCGUAU-ACGCAAUCUAUGAAAUAU-AAAUACAAAUCUGAAAAGCAGAAAGAGCGUCAGAAACUUUUCGAACCGUACAAAUAAAAUAA--------------------------------- ..((((((...-.))))))...........-.........(((((...((.......)).)))))............................--------------------------------- ( -13.50, z-score = -1.55, R) >droSec1.super_5 2048573 80 - 5866729 ACAUUGCGUAUAACGCAAUCUAUAAUAUAU-AAAUACAAAUCUGAAAAGCA------------GAAACUUUUCGAACCGGUCAAAUAAAAUAA--------------------------------- ..((((((.....))))))...........-...........(((((((..------------....)))))))...................--------------------------------- ( -10.80, z-score = -1.88, R) >droEre2.scaffold_4929 4003554 92 - 26641161 ACGUUGCGUAU-ACGCAACCUAUGAAGUAUGAAAAACAAGUUGGAAAAGCAGAAAGGGCGUCUGAAACUUUUCGAACCGUUCAAAUAAAAUAA--------------------------------- ..((((((...-.))))))..........((((......(((.((((((((((.......))))...)))))).)))..))))..........--------------------------------- ( -19.60, z-score = -1.46, R) >droAna3.scaffold_12943 492778 117 + 5039921 ACAUUGCGUAU-ACGCCAGGGUG--------GCUCCCACUUUUCGAAAAAGGGAACUGAGUGCGGAUUGAAUCCGAAAGUCCUUGGGUGGAAGUGGUGAACUUUUCAAACUGUUUGAAUAAAGUGC .(((..(...(-((.((((((..--------((((((.((((.....))))))....)))).(((((...))))).....)))))))))...)..))).(((((((((.....))))..))))).. ( -31.30, z-score = -0.35, R) >dp4.chr4_group3 9621211 70 + 11692001 ACAUUGCGUAU-ACGCAACGUU----------CGCCCGCUUU---AAAAGGGAAAACACUUCUAGAACACUUCGGUUAAUACGC------------------------------------------ .....((((((-.......(((----------..(((.....---....)))..)))...........((....))..))))))------------------------------------------ ( -14.80, z-score = -1.23, R) >droPer1.super_1 6736415 70 + 10282868 ACAUUGCGUAU-ACGCAGCGUU----------CGCCCGCUUU---AAAAGGGAAAACACUUCUAGAACACUUCGGUUAAUACGC------------------------------------------ .....((((((-..(.((.(((----------..(((.....---....)))..))).)).)......((....))..))))))------------------------------------------ ( -15.70, z-score = -1.00, R) >consensus ACAUUGCGUAU_ACGCAACCUAU_A__UAU_AAACACAAAUUUGAAAAGCAGAAAGCGCGUCUGAAACUUUUCGAACCGUACAAAUAAAAUAA_________________________________ ...(((((.....)))))............................................................................................................ ( -4.98 = -5.00 + 0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:02 2011