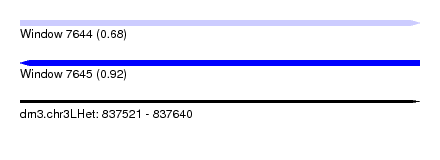
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 837,521 – 837,640 |
| Length | 119 |
| Max. P | 0.924779 |

| Location | 837,521 – 837,640 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.23 |
| Shannon entropy | 0.55258 |
| G+C content | 0.47779 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -10.91 |
| Energy contribution | -13.47 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
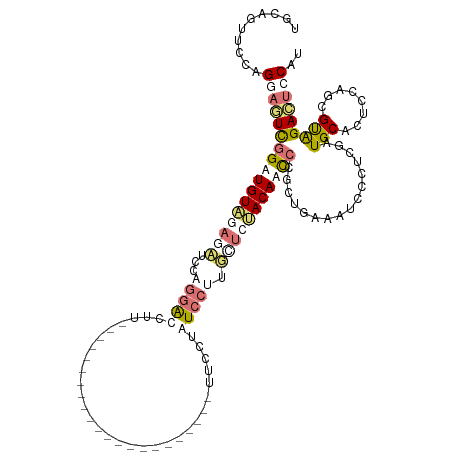
>dm3.chr3LHet 837521 119 + 2555491 ACUAACCCCAGUAGUCGGAUGUGAAGCUCUUAAACCUUAAACCUCUUAAAGAAGAAAAACUUCUUCUCCUUGUUCCACACUACGCUGUAAUCCUCCGAUGCUCUCCCACGCGGAGUCCA- .........((((.(((((.(...(((......................(((((((....)))))))....((........))))).....).)))))))))((((.....))))....- ( -20.90, z-score = -0.85, R) >droSim1.chr3R_random 152053 98 + 1307089 UGCAGUUCCAGGAGUUGGUUGUAGAUAUCCAGGAUCUU----------------------UUCCUAUCCUUUCUCUACAACCCUCUGAAAUCCCUCGGUGCACUUCAGCGUAGACUCCAU ..........(((((((((((((((.....(((((...----------------------.....)))))...)))))))))..((((......)))).((......))...)))))).. ( -27.50, z-score = -2.40, R) >droSec1.super_113 21515 98 + 77858 UGCAGUUCCAGGAGUUGGUUGUAGAUAUCCAGGAUCUU----------------------UUCCUAUCCUUGCUCUACAACCCUCUGAAAUCCCUCGGUGCACUUCAGCGUAGACUCCAU ..........(((((((((((((((...(.(((((...----------------------.....))))).).)))))))))..((((......)))).((......))...)))))).. ( -27.70, z-score = -2.07, R) >droEre2.scaffold_4845 21204513 98 - 22589142 UUCAGUUCUAGCAUUCGGAUGUAGUGCUCCUGGGCUUU----------------------UUCGUCUCCUUGCUAGACACUCUGCUGUACUCCCGUAAUGCACUCUAGCGUGGAGCUCAU ..(((.(((((((...(((((....(((....)))...----------------------..)))))...)))))))....))).((..(((((((...........))).))))..)). ( -27.70, z-score = -1.00, R) >consensus UGCAGUUCCAGGAGUCGGAUGUAGAGAUCCAGGACCUU______________________UUCCUAUCCUUGCUCUACAACCCGCUGAAAUCCCUCGAUGCACUCCAGCGUAGACUCCAU ..........(((((((((((((((((...((((................................)))).)))))))))))................(((........))))))))).. (-10.91 = -13.47 + 2.56)

| Location | 837,521 – 837,640 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.23 |
| Shannon entropy | 0.55258 |
| G+C content | 0.47779 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -14.21 |
| Energy contribution | -15.58 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
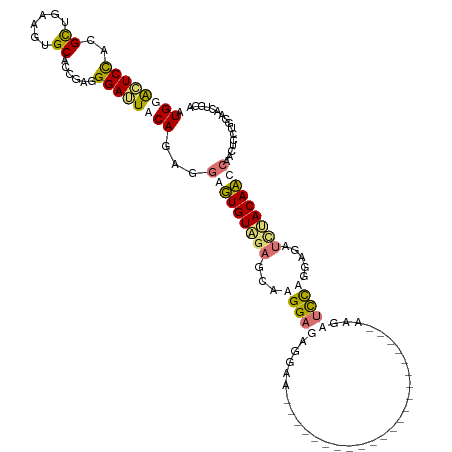
>dm3.chr3LHet 837521 119 - 2555491 -UGGACUCCGCGUGGGAGAGCAUCGGAGGAUUACAGCGUAGUGUGGAACAAGGAGAAGAAGUUUUUCUUCUUUAAGAGGUUUAAGGUUUAAGAGCUUCACAUCCGACUACUGGGGUUAGU -....(((((.(((......))))))))(((..((..(((((.((((....(((((((((....)))))))))..(((((((.........)))))))...)))))))))))..)))... ( -33.40, z-score = -0.85, R) >droSim1.chr3R_random 152053 98 - 1307089 AUGGAGUCUACGCUGAAGUGCACCGAGGGAUUUCAGAGGGUUGUAGAGAAAGGAUAGGAA----------------------AAGAUCCUGGAUAUCUACAACCAACUCCUGGAACUGCA ..(((((...(.(((((((.(......).))))))).)(((((((((...(((((.....----------------------...))))).....))))))))).))))).......... ( -34.50, z-score = -3.14, R) >droSec1.super_113 21515 98 - 77858 AUGGAGUCUACGCUGAAGUGCACCGAGGGAUUUCAGAGGGUUGUAGAGCAAGGAUAGGAA----------------------AAGAUCCUGGAUAUCUACAACCAACUCCUGGAACUGCA ..(((((...(.(((((((.(......).))))))).)(((((((((.(.(((((.....----------------------...))))).)...))))))))).))))).......... ( -34.70, z-score = -2.90, R) >droEre2.scaffold_4845 21204513 98 + 22589142 AUGAGCUCCACGCUAGAGUGCAUUACGGGAGUACAGCAGAGUGUCUAGCAAGGAGACGAA----------------------AAAGCCCAGGAGCACUACAUCCGAAUGCUAGAACUGAA ....(((((..((((((.(((..(((....)))..))).....))))))..(....)...----------------------........))))).((((((....))).)))....... ( -26.00, z-score = -0.85, R) >consensus AUGGACUCCACGCUGAAGUGCACCGAGGGAUUACAGAGGAGUGUAGAGCAAGGAGAGGAA______________________AAGAUCCAGGAGAUCUACAACCAACUCCUGGAACUGCA .((((((((..((......))......))))))))...(((((((((...(((((..............................))))).....)))))))))................ (-14.21 = -15.58 + 1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:54 2011