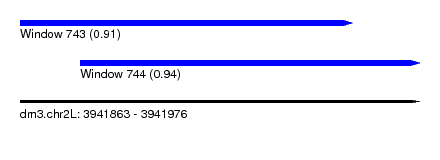
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,941,863 – 3,941,976 |
| Length | 113 |
| Max. P | 0.938234 |

| Location | 3,941,863 – 3,941,957 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Shannon entropy | 0.41646 |
| G+C content | 0.59988 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -26.18 |
| Energy contribution | -25.42 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.907914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
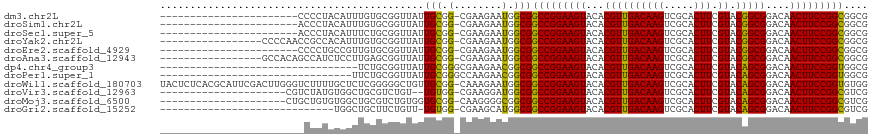
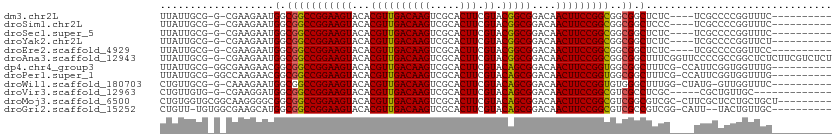
>dm3.chr2L 3941863 94 + 23011544 -----------------------CCCCUACAUUUGUGCGGUUAUUGCGG-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCG -----------------------......(((((.(((.((....)).)-))..)))))((.(((((((((...((((((((((.....))).)).)))))....))))))))).)). ( -37.30, z-score = -1.69, R) >droSim1.chr2L 3896773 94 + 22036055 -----------------------ACCCUACAUUUGUGCGGUUAUUGCGG-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCG -----------------------......(((((.(((.((....)).)-))..)))))((.(((((((((...((((((((((.....))).)).)))))....))))))))).)). ( -37.30, z-score = -1.87, R) >droSec1.super_5 2044726 94 + 5866729 -----------------------ACCCUACAUUUCUGCGGUUAUUGCGG-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCG -----------------------......(((((.(((.((....)).)-))..)))))((.(((((((((...((((((((((.....))).)).)))))....))))))))).)). ( -36.70, z-score = -2.00, R) >droYak2.chr2L 3956096 100 + 22324452 -----------------CCCCAACCGCCACAUUUGUGCGGUUAUUGCGG-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCG -----------------..((...(((((..(((.(((.((....)).)-)).))).)))))(((((((((...((((((((((.....))).)).)))))....))))))))))).. ( -39.30, z-score = -1.34, R) >droEre2.scaffold_4929 3999652 94 + 26641161 -----------------------CCCCUGCCGUUGUGCGGUUAUUGCGG-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCG -----------------------..(((((((((.(((.((....)).)-))...)))))))(((((((((...((((((((((.....))).)).)))))....))))))))))).. ( -38.60, z-score = -0.96, R) >droAna3.scaffold_12943 488732 100 - 5039921 -----------------GCCACAGCCAUCUCCUUGAGCGGUUAUUGCGG-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCG -----------------(((...(((((...(((..((.((....)).)-).))).))))).(((((((((...((((((((((.....))).)).)))))....)))))))))))). ( -42.70, z-score = -2.65, R) >dp4.chr4_group3 2076409 85 + 11692001 ---------------------------------UCUGCGGUUAUUGCGGGCGAAGAACGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGUGGCG ---------------------------------((((((.....)))))).........((.(((((((((...((((((((((.....))).)).)))))....))))))))).)). ( -33.10, z-score = -1.70, R) >droPer1.super_1 3552198 86 + 10282868 --------------------------------UUCUGCGGUUAUUGCGGGCCAAGAACGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGUGGCG --------------------------------((((..((((......)))).))))..((.(((((((((...((((((((((.....))).)).)))))....))))))))).)). ( -34.40, z-score = -2.14, R) >droWil1.scaffold_180703 1881476 117 + 3946847 UACUCUCACGCAUUCGACUUGGGUCUUUUGCUCUCGGGGGCUGUUGCGG-CAAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGUGUGG ......(((((((((..((((((.........)))))).((((...)))-)...)))).)).(((((((((...((((((((((.....))).)).)))))....)))))))))))). ( -39.00, z-score = -0.64, R) >droVir3.scaffold_12963 10867741 94 - 20206255 ---------------------CGUCUAUGUGGCUGCGUCUGU--UGUGG-CGAAGGAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGUCG ---------------------.((((...(.((..((.....--))..)-).).))))((((.((((((((...((((((((((.....))).)).)))))....)))))))))))). ( -37.10, z-score = -1.23, R) >droMoj3.scaffold_6500 1945986 96 + 32352404 ---------------------CUGCUGUGUGGCUGCGUCUGUGGUGCGG-CAAGGGGCGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGUCG ---------------------..(((.(...(((((..(...)..))))-)..).)))((((.((((((((...((((((((((.....))).)).)))))....)))))))))))). ( -42.10, z-score = -1.14, R) >droGri2.scaffold_15252 12469282 87 + 17193109 -----------------------------UGGCUGCUUCUGUU-UGUGG-CGAAGCAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGUCG -----------------------------....((((((.((.-....)-))))))).((((.((((((((...((((((((((.....))).)).)))))....)))))))))))). ( -35.20, z-score = -1.88, R) >consensus ________________________CCCUACAUUUGUGCGGUUAUUGCGG_CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGGCG ............................................(((.(........).)))(((((((((...((((((((((.....))).)).)))))....))))))))).... (-26.18 = -25.42 + -0.76)
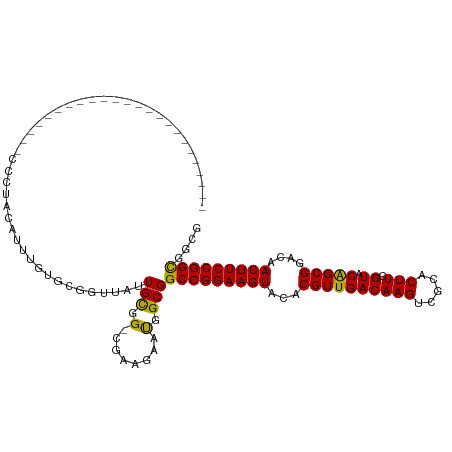
| Location | 3,941,880 – 3,941,976 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.27 |
| Shannon entropy | 0.35402 |
| G+C content | 0.60599 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -25.70 |
| Energy contribution | -25.35 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.938234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3941880 96 + 23011544 UUAUUGCG-G-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCGGCUCUC----UCGCCCCGGUUUC---------- ..((((.(-(-((((((.(.((.(((((((((...((((((((((.....))).)).)))))....))))))))).)).).))).----))))).))))...---------- ( -42.70, z-score = -2.61, R) >droSim1.chr2L 3896790 96 + 22036055 UUAUUGCG-G-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCGGCUCCC----UCGCCCCGGUUUC---------- ..((((.(-(-(((.((.(.((.(((((((((...((((((((((.....))).)).)))))....))))))))).)).).))..----))))).))))...---------- ( -41.00, z-score = -1.75, R) >droSec1.super_5 2044743 96 + 5866729 UUAUUGCG-G-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCGGCUCUC----UCGCCCCGGUUUC---------- ..((((.(-(-((((((.(.((.(((((((((...((((((((((.....))).)).)))))....))))))))).)).).))).----))))).))))...---------- ( -42.70, z-score = -2.61, R) >droYak2.chr2L 3956119 96 + 22324452 UUAUUGCG-G-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCGGCUCUC----UCGCCCCGGUUCU---------- ..((((.(-(-((((((.(.((.(((((((((...((((((((((.....))).)).)))))....))))))))).)).).))).----))))).))))...---------- ( -42.70, z-score = -2.52, R) >droEre2.scaffold_4929 3999669 96 + 26641161 UUAUUGCG-G-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCGGCUCUC----UCGCCCCGGUUCC---------- ..((((.(-(-((((((.(.((.(((((((((...((((((((((.....))).)).)))))....))))))))).)).).))).----))))).))))...---------- ( -42.70, z-score = -2.36, R) >droAna3.scaffold_12943 488755 110 - 5039921 UUAUUGCG-G-CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACGGCGGACAACUUCCGGCGGCGGCUUUCGGUUCCCCGCCGGCUCUCUUCGUCUCU .......(-(-((((((..(((.(((((((((...((((((((((.....))).)).)))))....)))))))))(((((...........))))).))).))))))))... ( -47.70, z-score = -2.37, R) >dp4.chr4_group3 2076416 100 + 11692001 UUAUUGCG-GGCGAAGAACGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGUGGCGGCUUUCG-CCAUUCGGUGGUUUG---------- ((((((..-(((((((..((.(.(((((((((...((((((((((.....))).)).)))))....))))))))).)))..)))))-))...))))))....---------- ( -42.50, z-score = -2.99, R) >droPer1.super_1 3552206 100 + 10282868 UUAUUGCG-GGCCAAGAACGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGUGGCGGCUUUCG-CCAUUCGGUGGUUUG---------- ......((-(((((.(((..((.(((((((((...((((((((((.....))).)).)))))....))))))))).))(((....)-)).)))..)))))))---------- ( -41.70, z-score = -2.77, R) >droWil1.scaffold_180703 1881516 98 + 3946847 CUGUUGCG-G-CAAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGUGUGGGCUUUGG-CUAUG-GUUGGUUUC---------- .......(-.-(((((..(.(((.((((((((...((((((((((.....))).)).)))))....))))))))))).).))))).-)....-.........---------- ( -30.40, z-score = -0.03, R) >droVir3.scaffold_12963 10867758 92 - 20206255 CUGUUGUG-G-CGAAGGAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGUCGCCUCGC-----CGCUGUUGC------------- .....(((-(-(((.((.(((((.((((((((...((((((((((.....))).)).)))))....)))))))))))))))))))-----)))......------------- ( -44.60, z-score = -3.24, R) >droMoj3.scaffold_6500 1946003 102 + 32352404 CUGUGGUGCGGCAAGGGGCGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGUCGGCGUCGC-CUUCGCUCCUGCUGCU--------- .......((((((.(((((((((.((((((((...((((((((((.....))).)).)))))....))))))))))))(((...))-)...))))))))))).--------- ( -50.40, z-score = -2.60, R) >droGri2.scaffold_15252 12469291 98 + 17193109 CUGUU-UGUGGCGAAGCAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGUCGCCGUCGG-CAUU--UACUGUUGC---------- ..((.-.(((.(((....((((((((((((((...((((((((((.....))).)).)))))....)))))))))..)))))))).-))).--.))......---------- ( -39.50, z-score = -1.86, R) >consensus UUAUUGCG_G_CGAAGAAUGGCGGCCGGAAGUACACGUUGACAAGUCGCACUUCGUACAGCGGACAACUUCCGGCGGCGGCUCUC____UCGCCCCGGUUUC__________ ...................(.(((((((((((...((((((((((.....))).)).)))))....)))))))))..)).)............................... (-25.70 = -25.35 + -0.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:01 2011