
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 665,542 – 665,673 |
| Length | 131 |
| Max. P | 0.614182 |

| Location | 665,542 – 665,673 |
|---|---|
| Length | 131 |
| Sequences | 3 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 57.51 |
| Shannon entropy | 0.61542 |
| G+C content | 0.56754 |
| Mean single sequence MFE | -46.63 |
| Consensus MFE | -16.06 |
| Energy contribution | -17.20 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614182 |
| Prediction | RNA |
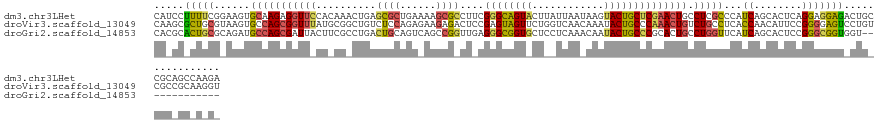
Download alignment: ClustalW | MAF
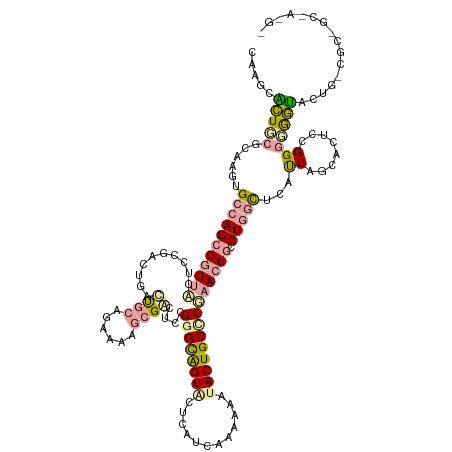
>dm3.chr3LHet 665542 131 + 2555491 CAUCCUUUUCGGAAGUGCAAGAGGUUCCACAAACUGAGCGCUGAAAAGCGCCUUCGGGCAGUACUUAUUAAUAAGUACUGCUCGAACUGCCUCGCCCAUCAGCACUCAGGAGGAGACUGCCGCAGCCAAGA ..........((...(((..(((((............(((((....))))).(((((((((((((((....)))))))))))))))..)))))((......)).....((((....)).))))).)).... ( -50.90, z-score = -3.65, R) >droVir3.scaffold_13049 24538204 131 + 25233164 CAAGCGCUGCGUAAGUGCCAGCGGUUUAUGCGGCUGUCUCCAGAGAAGAGACUCCGAGUAGUUCUGGUCAACAAAUACUGCCCAAACUGUCUGCCUCACCAACAUUCCGGGGAGUCCUGUCGCCGCAAGGU .((.(((((.((....))))))).))..((((((..(((....)))((.((((((((((((((.(((.((........)).)))))))).....))).((........))))))))))...)))))).... ( -41.30, z-score = -0.28, R) >droGri2.scaffold_14853 8980702 118 - 10151454 CACGCACUGCGCAGAUGCCAGCGAUUACUUCGCCUGACUGCAGUCAGCCGGUUGAGGGCGGUGCUCCUCAAACAAUACUGCCCGCACUGCCUGGUUCAUCAGCACUCCGGGCGGUGGU------------- ...(((((((.(....(((.((((.....))))(((((....)))))..)))...).)))))))...........(((((((((...(((.(((....))))))...)))))))))..------------- ( -47.70, z-score = -1.07, R) >consensus CAAGCACUGCGCAAGUGCCAGCGGUUACUCCGACUGACUGCAGAAAAGCGACUCCGGGCAGUACUCAUCAAAAAAUACUGCCCGAACUGCCUGGCUCAUCAGCACUCCGGGGGGUACUG_CGC_GC_A_G_ .....(((((......(((((((((((..........((((......))))....((((((((............)))))))))))))).)))))...((........)))))))................ (-16.06 = -17.20 + 1.14)
| Location | 665,542 – 665,673 |
|---|---|
| Length | 131 |
| Sequences | 3 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 57.51 |
| Shannon entropy | 0.61542 |
| G+C content | 0.56754 |
| Mean single sequence MFE | -49.37 |
| Consensus MFE | -16.88 |
| Energy contribution | -15.33 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
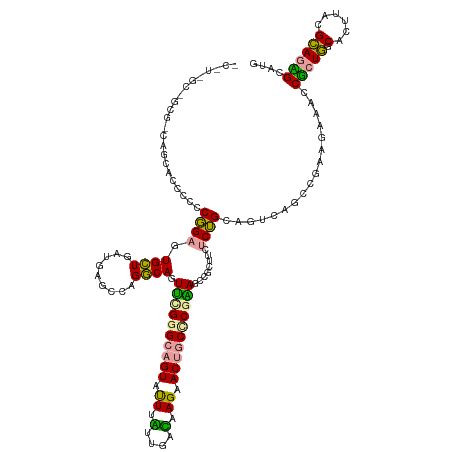
>dm3.chr3LHet 665542 131 - 2555491 UCUUGGCUGCGGCAGUCUCCUCCUGAGUGCUGAUGGGCGAGGCAGUUCGAGCAGUACUUAUUAAUAAGUACUGCCCGAAGGCGCUUUUCAGCGCUCAGUUUGUGGAACCUCUUGCACUUCCGAAAAGGAUG ..((((.(((((.((..(((.((((((((((((..(((...((..((((.((((((((((....)))))))))).)))).)))))..)))))))))))...).)))..)).)))))...))))........ ( -56.60, z-score = -3.84, R) >droVir3.scaffold_13049 24538204 131 - 25233164 ACCUUGCGGCGACAGGACUCCCCGGAAUGUUGGUGAGGCAGACAGUUUGGGCAGUAUUUGUUGACCAGAACUACUCGGAGUCUCUUCUCUGGAGACAGCCGCAUAAACCGCUGGCACUUACGCAGCGCUUG ....(((((((((((((((.((((((.((((.(.....).)))).)))))).))).))))))..((((.....)).)).((((((.....)))))).)))))).....(((((.(......)))))).... ( -44.40, z-score = -0.56, R) >droGri2.scaffold_14853 8980702 118 + 10151454 -------------ACCACCGCCCGGAGUGCUGAUGAACCAGGCAGUGCGGGCAGUAUUGUUUGAGGAGCACCGCCCUCAACCGGCUGACUGCAGUCAGGCGAAGUAAUCGCUGGCAUCUGCGCAGUGCGUG -------------..(((.(((((.(.((((.........)))).).))))).(((((((((((((.((...))))))))(((((((....))))).(((((.....))))))).......)))))))))) ( -47.10, z-score = -0.51, R) >consensus _C_U_GC_GCG_CAGCACCCCCCGGAGUGCUGAUGAGCCAGGCAGUUCGGGCAGUAUUUAUUGACAAGAACUGCCCGAAGCCGCUUCUCUGCAGUCAGCCGAAGAAACCGCUGGCACUUACGCAGAGCAUG ......................((((.((((.........)))).((((((((((.((.(....).)).))))))))))........)))).................(((((.(......)))))).... (-16.88 = -15.33 + -1.54)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:48 2011