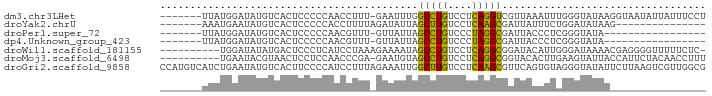
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 660,478 – 660,561 |
| Length | 83 |
| Max. P | 0.908884 |

| Location | 660,478 – 660,561 |
|---|---|
| Length | 83 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 58.33 |
| Shannon entropy | 0.79055 |
| G+C content | 0.43062 |
| Mean single sequence MFE | -16.35 |
| Consensus MFE | -7.57 |
| Energy contribution | -7.23 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.34 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
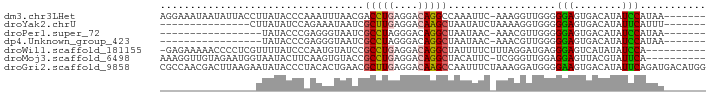
>dm3.chr3LHet 660478 83 + 2555491 AGGAAAUAAUAUUACCUUAUACCCAAAUUUAACGACCUGAGGACAGGCCAAAUUC-AAAGGUUGGGGGAGUGACAUAUCCAUAA------- .(((.((..((((.(((((.(((..........(.((((....))))).......-...)))))))).))))..)).)))....------- ( -14.95, z-score = 0.44, R) >droYak2.chrU 11499327 69 + 28119190 ---------------CUUAUAUCCAGAAAUAAUCGCUUGAGGACAAGCUAAUAUCUAAAAGGUGGGGGAGUGACAUAUUCAUUU------- ---------------......(((..........(((((....)))))...((((.....)))))))((((((.....))))))------- ( -10.90, z-score = -0.02, R) >droPer1.super_72 265259 66 - 283643 -----------------UAUACCCGAGGGUAAUCGCCUAGGGACAGGCUAAUAAC-AAACGUUGGGGGAGUGACAUAUCCAUAA------- -----------------....(((((.((....)((((......)))).......-...).)))))(((........)))....------- ( -15.00, z-score = -0.08, R) >dp4.Unknown_group_423 15081 66 - 54704 -----------------UAUACCCGAGGGUAAUCGCCUAGGGACAGGCUAAUAAC-AAACGUUGGGGGAGUGACAUAUCCAUAA------- -----------------....(((((.((....)((((......)))).......-...).)))))(((........)))....------- ( -15.00, z-score = -0.08, R) >droWil1.scaffold_181155 1907240 80 + 3442787 -GAGAAAAACCCCUCGUUUUAUCCCAAUGUAUCCGCCUGAGGACAGGCUAUUUUCUUUAGGAUGAGGGAGUCAUAUAUCCA---------- -.......(((((((((((((.......(((...(((((....))))))))......))))))))))).))..........---------- ( -21.32, z-score = -1.29, R) >droMoj3.scaffold_6498 3143416 80 + 3408170 AAAGGUUGUAGAAUGGUAAUACUUCAAGUGUACCGCCUGAGGACAGGCUACAUUC-UCGGGUUGGAGGAGUUACGUAUUCA---------- ..........((((((((((.(((((((((((..(((((....))))))))))..-.....))))))..))))).))))).---------- ( -22.01, z-score = -1.17, R) >droGri2.scaffold_9858 1266 91 + 5805 CGCCAACGACUUAAGAAUAUACCCUACACUGAACGCUUGAGGACAAGCCAAUUUCUAAAGGAUGGGGAAGUGACAUAUUCAGAUGACAUGG ..((.................(((((..(((((.(((((....)))))....)))...))..)))))..(((.(((......))).))))) ( -15.30, z-score = -0.19, R) >consensus ________________UUAUACCCAAAGGUAACCGCCUGAGGACAGGCUAAUUUC_AAAGGUUGGGGGAGUGACAUAUCCAUAA_______ ..................................(((((....)))))..................(((........)))........... ( -7.57 = -7.23 + -0.35)
| Location | 660,478 – 660,561 |
|---|---|
| Length | 83 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 58.33 |
| Shannon entropy | 0.79055 |
| G+C content | 0.43062 |
| Mean single sequence MFE | -15.85 |
| Consensus MFE | -7.66 |
| Energy contribution | -6.93 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
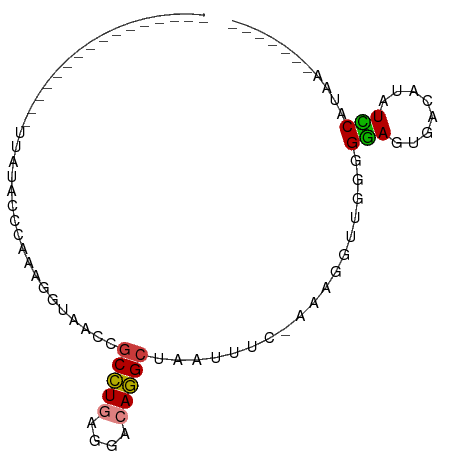
>dm3.chr3LHet 660478 83 - 2555491 -------UUAUGGAUAUGUCACUCCCCCAACCUUU-GAAUUUGGCCUGUCCUCAGGUCGUUAAAUUUGGGUAUAAGGUAAUAUUAUUUCCU -------....(((.((((.(((..(((((..(((-((...(((((((....)))))))))))).))))).....))).))))....))). ( -19.90, z-score = -1.29, R) >droYak2.chrU 11499327 69 - 28119190 -------AAAUGAAUAUGUCACUCCCCCACCUUUUAGAUAUUAGCUUGUCCUCAAGCGAUUAUUUCUGGAUAUAAG--------------- -------.......((((((.......................(((((....)))))((.....))..))))))..--------------- ( -10.60, z-score = -1.37, R) >droPer1.super_72 265259 66 + 283643 -------UUAUGGAUAUGUCACUCCCCCAACGUUU-GUUAUUAGCCUGUCCCUAGGCGAUUACCCUCGGGUAUA----------------- -------....(((........)))..........-.......(((((....)))))...((((....))))..----------------- ( -12.00, z-score = 0.52, R) >dp4.Unknown_group_423 15081 66 + 54704 -------UUAUGGAUAUGUCACUCCCCCAACGUUU-GUUAUUAGCCUGUCCCUAGGCGAUUACCCUCGGGUAUA----------------- -------....(((........)))..........-.......(((((....)))))...((((....))))..----------------- ( -12.00, z-score = 0.52, R) >droWil1.scaffold_181155 1907240 80 - 3442787 ----------UGGAUAUAUGACUCCCUCAUCCUAAAGAAAAUAGCCUGUCCUCAGGCGGAUACAUUGGGAUAAAACGAGGGGUUUUUCUC- ----------.(((.....(((.((((((((((((........(((((....))))).......))))))).....))))))))..))).- ( -24.76, z-score = -2.02, R) >droMoj3.scaffold_6498 3143416 80 - 3408170 ----------UGAAUACGUAACUCCUCCAACCCGA-GAAUGUAGCCUGUCCUCAGGCGGUACACUUGAAGUAUUACCAUUCUACAACCUUU ----------........................(-(((((..(((((....)))))(((((.......)))))..))))))......... ( -15.70, z-score = -1.37, R) >droGri2.scaffold_9858 1266 91 - 5805 CCAUGUCAUCUGAAUAUGUCACUUCCCCAUCCUUUAGAAAUUGGCUUGUCCUCAAGCGUUCAGUGUAGGGUAUAUUCUUAAGUCGUUGGCG (((((.(....(((((((......(((.(..((...(((....(((((....))))).)))))..).))))))))))....).)).))).. ( -16.00, z-score = -0.10, R) >consensus _______UUAUGGAUAUGUCACUCCCCCAACCUUU_GAAAUUAGCCUGUCCUCAGGCGAUUACCUUCGGGUAUAA________________ ...........................................(((((....))))).................................. ( -7.66 = -6.93 + -0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:47 2011