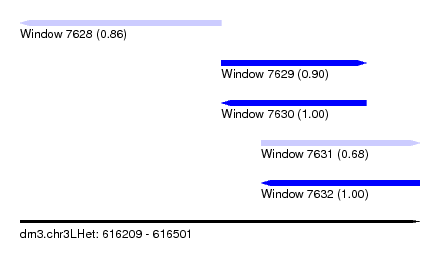
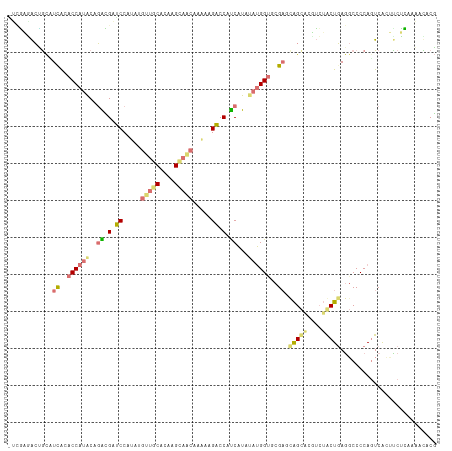
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 616,209 – 616,501 |
| Length | 292 |
| Max. P | 0.999619 |

| Location | 616,209 – 616,356 |
|---|---|
| Length | 147 |
| Sequences | 5 |
| Columns | 147 |
| Reading direction | reverse |
| Mean pairwise identity | 56.73 |
| Shannon entropy | 0.79827 |
| G+C content | 0.52020 |
| Mean single sequence MFE | -52.70 |
| Consensus MFE | -13.12 |
| Energy contribution | -17.60 |
| Covariance contribution | 4.48 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
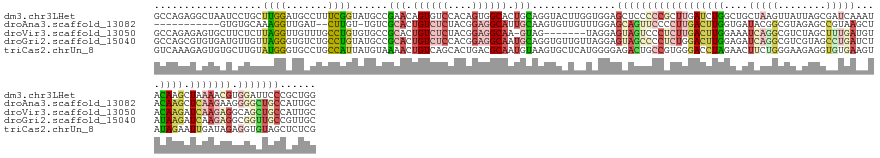
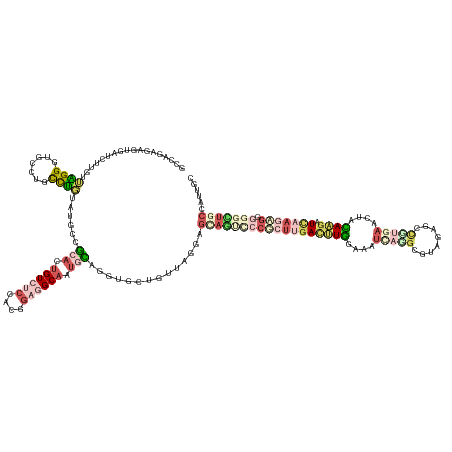
>dm3.chr3LHet 616209 147 - 2555491 GCCAGAGGCUAAUCCUGCUUGGAUGCCUUUCGGUAUGCCGAACAGUGUCCACAGUGGCACUGCAGGUACUUGGUGGAGCUCCCCCGCUUGAUCUGGCUGCUAAGUUAUUAGCGAUCAAAUACAAGCUAAAACGUGGAUUCCCGCUGG (((((((((..((((.....)))))))))).))).((((...(((((((......)))))))..))))...((.((....)).))(((((((.((..((((((....))))))..)).)).)))))......((((....))))... ( -53.10, z-score = -1.82, R) >droAna3.scaffold_13082 239205 133 - 3919855 -----------GUGUGCAAAGGUUGAU--CUUGU-UGUCGCACUGUCUCUACGGAGGCAUUGCAAGUGUUGUUUGGAGCAGUUCCCCUUGACUUGGUGAUACGGCGUAGAGCCGUAAGCUACAAGCUCAAGAAGGGGCUGCCAUUGC -----------(((((((((((.....--))).)-)).)))))((((((....))))))..((((..........(.((((((((.(((((((((....((((((.....)))))).....)))).)))))..))))))))).)))) ( -49.80, z-score = -1.84, R) >droVir3.scaffold_13050 451069 139 - 3426691 GCCAGAGAGUGCUUCUCUUAGGUUGUUUGCCUGUGUGCCGCACUGUCUCUACGGAGGCAA-GUAG-------UAGGAGUAGUCCCUCUUGACUUGGAAAUCAGGCGUCUAGCUUUGAUGUACAAGAUCAAGAGGCAGCUGCCAUUGC (((.((.(((((..(.(.(((((.....))))).).)..))))).))((....)))))..-...(-------(((..(((((.((((((((((((...(((((((.....)).)))))...)))).))))))))..)))))..)))) ( -54.80, z-score = -2.54, R) >droGri2.scaffold_15040 117516 147 - 162499 GCCAGCGUGUGAUGUUGUUAGGGUGUCUGCCUGUAUGCCGCACUGUCUCCACGGAGGCAAUGCAGGUGUUGUUAGGAGUAGCCCCUCUGGACUUGGAGAUCAGGCGUCGUAGCCUGAUCUAUAAGAUCAAGAGGCGGUUGCCGUUGC ..(((((...((..((.....))..))..((((.((((((((.((((((....)))))).))).)))))...)))).(((((((((((.((((((.(((((((((......))))))))).)))).)).))))).))))))))))). ( -65.30, z-score = -3.69, R) >triCas2.chrUn_8 580423 147 - 609019 GUCAAAGAGUGUGCUUGUAUGGGUGCCUGCCAUUAUGUAAAACUGUCAGCACUGACGCAAUGUAAGUGCUCAUGGGGAGACUGCCGUGGGACCUAGAACUUCUGGGAAGAGGUGUGAAGUAUAGAAUUGAUAGAGGUGUAGCUCUCG (((((....(((((((...(((((.((..(............((((.((((((.((.....)).)))))).))))(....)....)..)))))))..((((((....))))))...)))))))...))))).((((......)))). ( -40.50, z-score = 0.29, R) >consensus GCCAGAGAGUGAUCUUGUUAGGGUGCCUGCCUGUAUGCCGCACUGUCUCCACGGAGGCAAUGCAGGUGCUGUUAGGAGCAGUCCCGCUUGACUUGGAAAUCAGGCGUAGAGCCGUGAACUACAAGAUCAAGAGGGGGCUGCCAUUGC ..................((((.......))))......(((.((((((....)))))).)))..............((((((((((((((((((....(((((........)))))....)))).))))))).)))))))...... (-13.12 = -17.60 + 4.48)
| Location | 616,356 – 616,462 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 55.03 |
| Shannon entropy | 0.76841 |
| G+C content | 0.49379 |
| Mean single sequence MFE | -26.11 |
| Consensus MFE | -9.43 |
| Energy contribution | -11.80 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 616356 106 + 2555491 AGGAGGCUAGUCGCCUCUGGGCAGCUACUUGUUCCGCGGUGUCAUACCAUACCGGGGCACCAAACAGCACACAAGGUGCCAUGAGUCCGUCAUAUAUGGUCCGCCU .((((((.....))))))((((.((((..(((...((((..((((.((.....))((((((.............))))))))))..)))).)))..))))..)))) ( -38.12, z-score = -1.01, R) >droAna3.scaffold_13082 239338 103 + 3919855 ---UUAAAAUUUUCAUUCUACCCCUAACGUCCAAGACUGCAUCGUACCACGCAGGUGAUCCGCUUGUUGCGCAAGUAACAAAAAGACCACCAUAUAUGGUGCGAGC ---.........................(((...))).((.((((((((....((((.((...(((((((....)))))))...)).)))).....)))))))))) ( -29.80, z-score = -3.72, R) >droVir3.scaffold_13050 451208 106 + 3426691 AAGCCAGCACUUUCUUUCUGCAAACCACAAUCGUGAACGCCUUGCACCAUACAUACGAUCCAUAUGCUGCACAUGCAGCAAACAGACCCUCAUAGAUGGUACGAAC ..((..(((.........)))....(((....)))...)).(((.(((((...((.((((....((((((....))))))....))...)).)).))))).))).. ( -19.60, z-score = -0.77, R) >droGri2.scaffold_15040 117663 106 + 162499 AAGCCAGCACAUUCUUUCUGCCUACAACAGUCGAAACUACUUUACACCAUACCGACGAUCCAUAAGUUGCACAGGCAACAAAUAGACCAUCGUAUACGGCGCGAGC ..(((.(((.........))).(((....((((...................)))).........(((((....)))))............)))...)))...... ( -16.91, z-score = -0.18, R) >consensus AAGCCAGCACUUUCUUUCUGCCAACAACAUUCGAGACUGCAUCACACCAUACAGACGAUCCAUAUGUUGCACAAGCAACAAAAAGACCAUCAUAUAUGGUGCGAGC .........................................((((((((((..((.(.((....((((((....))))))....)).).))...)))))))))).. ( -9.43 = -11.80 + 2.37)

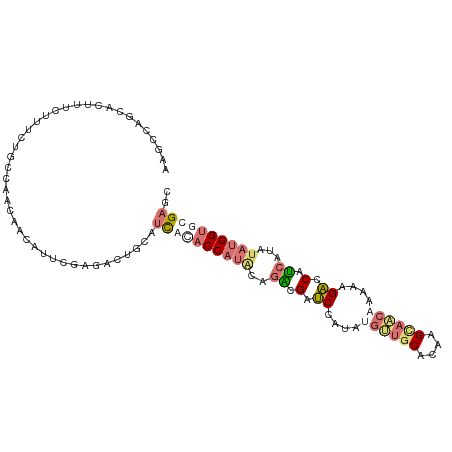
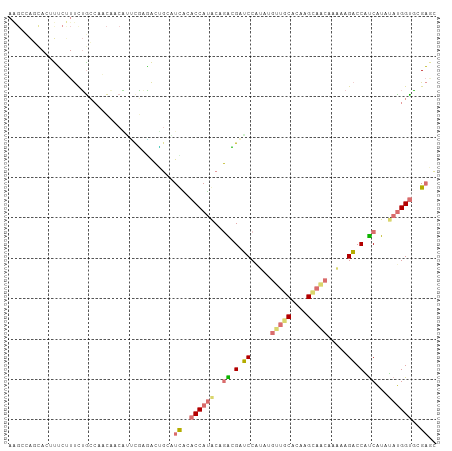
| Location | 616,356 – 616,462 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 55.03 |
| Shannon entropy | 0.76841 |
| G+C content | 0.49379 |
| Mean single sequence MFE | -36.91 |
| Consensus MFE | -20.33 |
| Energy contribution | -22.45 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 616356 106 - 2555491 AGGCGGACCAUAUAUGACGGACUCAUGGCACCUUGUGUGCUGUUUGGUGCCCCGGUAUGGUAUGACACCGCGGAACAAGUAGCUGCCCAGAGGCGACUAGCCUCCU .((((((((((((((((.....))))((((((.............))))))...)))))))........((.......))..)))))..(((((.....))))).. ( -34.92, z-score = 0.22, R) >droAna3.scaffold_13082 239338 103 - 3919855 GCUCGCACCAUAUAUGGUGGUCUUUUUGUUACUUGCGCAACAAGCGGAUCACCUGCGUGGUACGAUGCAGUCUUGGACGUUAGGGGUAGAAUGAAAAUUUUAA--- (((((.((((..((.((((((((.((((((........)))))).))))))))))..)))).))).)).(((...)))........((((((....)))))).--- ( -32.40, z-score = -1.95, R) >droVir3.scaffold_13050 451208 106 - 3426691 GUUCGUACCAUCUAUGAGGGUCUGUUUGCUGCAUGUGCAGCAUAUGGAUCGUAUGUAUGGUGCAAGGCGUUCACGAUUGUGGUUUGCAGAAAGAAAGUGCUGGCUU .((((((((((.((((..(((((((.((((((....)))))).))))))).)))).)))))))...(((.((((....))))..))).)))............... ( -36.50, z-score = -1.89, R) >droGri2.scaffold_15040 117663 106 - 162499 GCUCGCGCCGUAUACGAUGGUCUAUUUGUUGCCUGUGCAACUUAUGGAUCGUCGGUAUGGUGUAAAGUAGUUUCGACUGUUGUAGGCAGAAAGAAUGUGCUGGCUU (((.(((((((((.(((((((((((..(((((....)))))..))))))))))))))))))))..((((..(((..((((.....))))...)))..))))))).. ( -43.80, z-score = -4.95, R) >consensus GCUCGCACCAUAUAUGAUGGUCUAUUUGCUGCUUGUGCAACAUAUGGAUCGCCGGUAUGGUACAAAGCAGUCUCGAAUGUUGUUGGCAGAAAGAAAGUGCUGGCUU ....(((((((((.(((((((((((.((((((....)))))).))))))))))))))))))))........................................... (-20.33 = -22.45 + 2.12)

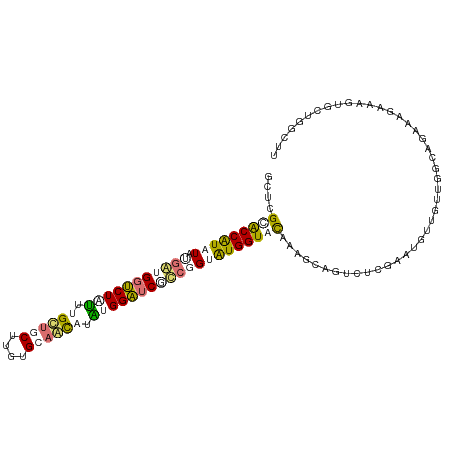
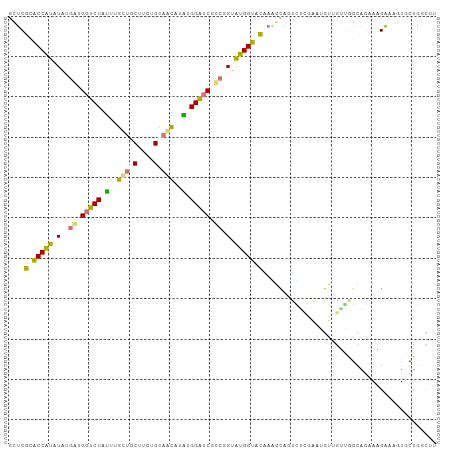
| Location | 616,385 – 616,501 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 59.77 |
| Shannon entropy | 0.67899 |
| G+C content | 0.51392 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -11.46 |
| Energy contribution | -13.77 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 616385 116 + 2555491 UGUUCCGCGGUGUCAUACCAUACCGGGGCACCAAACAGCACACAAGGUGCCAUGAGUCCGUCAUAUAUGGUCCGCCUGGCUCGAGGACUGAAGCCCCAGUCGGCUCGAAGCACACU ......((((......((((((....((((((.............))))))((((.....)))).))))))))))...((((((((((((......)))))..)))).)))..... ( -37.22, z-score = 0.01, R) >droAna3.scaffold_13082 239367 113 + 3919855 ---AAGACUGCAUCGUACCACGCAGGUGAUCCGCUUGUUGCGCAAGUAACAAAAAGACCACCAUAUAUGGUGCGAGCAGCUUUUCUGCUGAGGCCCCAUUCACUUCUUAAAACUCG ---(((.((((.((((((((....((((.((...(((((((....)))))))...)).)))).....))))))))))))))).......(((....................))). ( -37.15, z-score = -3.56, R) >droVir3.scaffold_13050 451237 116 + 3426691 AUCGUGAACGCCUUGCACCAUACAUACGAUCCAUAUGCUGCACAUGCAGCAAACAGACCCUCAUAGAUGGUACGAACAGUACGUCUACUUAGCCCCCAGUCAGACCUCAAAAUACG ..((((.((...(((.(((((...((.((((....((((((....))))))....))...)).)).))))).)))...))..(((((((........))).)))).......)))) ( -22.40, z-score = -1.14, R) >droGri2.scaffold_15040 117692 116 + 162499 GUCGAAACUACUUUACACCAUACCGACGAUCCAUAAGUUGCACAGGCAACAAAUAGACCAUCGUAUACGGCGCGAGCAGCAGUUCUGCUGAGGCCCCAGUCACUUCUCAAAAUCCG ((((...................)))).........(((((....))))).....(((....(....)((.((...(((((....)))))..)).)).)))............... ( -22.81, z-score = -0.32, R) >consensus _UCGAGACUGCAUCACACCAUACAGACGAUCCAUAUGUUGCACAAGCAACAAAAAGACCAUCAUAUAUGGUGCGAGCAGCACGUCUACUGAGGCCCCAGUCACUUCUCAAAACACG ............((((((((((..((.(.((....((((((....))))))....)).).))...)))))))))).(((((....))))).......................... (-11.46 = -13.77 + 2.31)


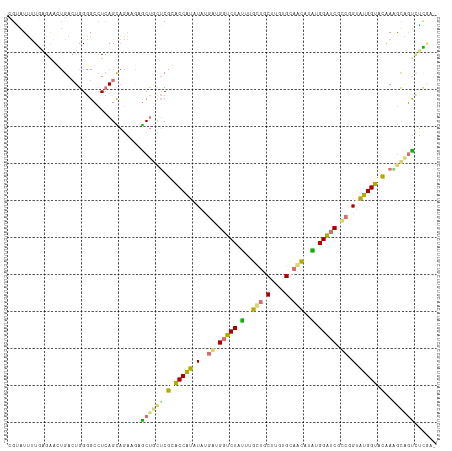
| Location | 616,385 – 616,501 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 59.77 |
| Shannon entropy | 0.67899 |
| G+C content | 0.51392 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -23.76 |
| Energy contribution | -26.57 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 616385 116 - 2555491 AGUGUGCUUCGAGCCGACUGGGGCUUCAGUCCUCGAGCCAGGCGGACCAUAUAUGACGGACUCAUGGCACCUUGUGUGCUGUUUGGUGCCCCGGUAUGGUAUGACACCGCGGAACA .(((.(..(((.((((((((((((.((((((((((.....((....)).....))).))))..(((((((.....))))))).))).)))))))).)))).)))..))))...... ( -40.90, z-score = 0.32, R) >droAna3.scaffold_13082 239367 113 - 3919855 CGAGUUUUAAGAAGUGAAUGGGGCCUCAGCAGAAAAGCUGCUCGCACCAUAUAUGGUGGUCUUUUUGUUACUUGCGCAACAAGCGGAUCACCUGCGUGGUACGAUGCAGUCUU--- .((((((((.........))))).))).........((((((((.((((..((.((((((((.((((((........)))))).))))))))))..)))).))).)))))...--- ( -35.10, z-score = -0.87, R) >droVir3.scaffold_13050 451237 116 - 3426691 CGUAUUUUGAGGUCUGACUGGGGGCUAAGUAGACGUACUGUUCGUACCAUCUAUGAGGGUCUGUUUGCUGCAUGUGCAGCAUAUGGAUCGUAUGUAUGGUGCAAGGCGUUCACGAU ..........(((((......)))))..((..((((.((....(((((((.((((..(((((((.((((((....)))))).))))))).)))).))))))).))))))..))... ( -40.00, z-score = -1.83, R) >droGri2.scaffold_15040 117692 116 - 162499 CGGAUUUUGAGAAGUGACUGGGGCCUCAGCAGAACUGCUGCUCGCGCCGUAUACGAUGGUCUAUUUGUUGCCUGUGCAACUUAUGGAUCGUCGGUAUGGUGUAAAGUAGUUUCGAC ......((((((....(((.((((...((((....))))))))(((((((((.(((((((((((..(((((....)))))..))))))))))))))))))))..)))..)))))). ( -47.40, z-score = -4.27, R) >consensus CGUAUUUUGAGAACUGACUGGGGCCUCAGCAGAAGAGCUGCUCGCACCAUAUAUGAUGGUCUAUUUGCUGCUUGUGCAACAUAUGGAUCGCCGGUAUGGUACAAAGCAGUCUCGA_ ....................................((((((((((((((((.(((((((((((.((((((....)))))).))))))))))))))))))))).))))))...... (-23.76 = -26.57 + 2.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:44 2011