
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,937,846 – 3,937,939 |
| Length | 93 |
| Max. P | 0.511409 |

| Location | 3,937,846 – 3,937,939 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Shannon entropy | 0.37262 |
| G+C content | 0.44169 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -8.99 |
| Energy contribution | -8.91 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
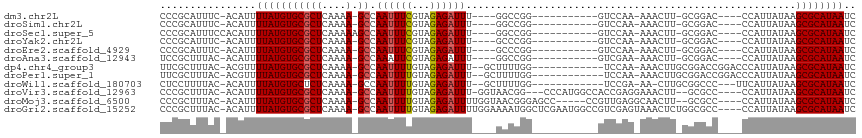
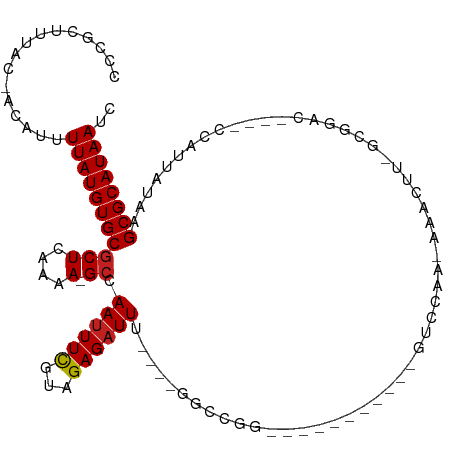
>dm3.chr2L 3937846 93 + 23011544 CCCGCAUUUC-ACAUUUUAUGUGCGCUCAAAA-GCCAAUUUCGUAGAGAUUU----GGCCGG-----------GUCCAA-AAACUU-GCGGAC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((.....-(((((..((.....)).))----))).((-----------((((..-......-..))))----))......))))))))... ( -30.20, z-score = -3.89, R) >droSim1.chr2L 3892828 93 + 22036055 CCCGCAUUUC-ACAUUUUAUGUGCGCUCAAAA-GCCAAUUUCGUAGAGAUUU----GGCCGG-----------GUCCAA-AAACUU-GCGGAC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((.....-(((((..((.....)).))----))).((-----------((((..-......-..))))----))......))))))))... ( -30.20, z-score = -3.89, R) >droSec1.super_5 2040812 95 + 5866729 CCCGCAUUUCCACAUUUUAUGUGCGCUCAAAAAGCCAAUUUCGUAGAGAUUU----GGCCGG-----------GUCCAA-AAACUU-GCGGAC----CCAUUAUAAGCGCAUAAUC ...................((((((((......(((((..((.....)).))----))).((-----------((((..-......-..))))----))......))))))))... ( -30.20, z-score = -3.76, R) >droYak2.chr2L 3952072 93 + 22324452 CCCGCAUUUC-ACAUUUUAUGUGCGCUCAAAA-GCCAAUUUCGUAGAGAUUU----GCCCGG-----------GUCCAA-AAACUU-GCGGAC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((.....-((.((((((...)))))).----))..((-----------((((..-......-..))))----))......))))))))... ( -27.10, z-score = -3.36, R) >droEre2.scaffold_4929 3995608 93 + 26641161 CCCGCAUUUC-ACAUUUUAUGUGCGCUCAAAA-GCCAAUUUCGUAGAGAUUU----GCCCGG-----------GUCCAA-AAACUU-GCGGAC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((.....-((.((((((...)))))).----))..((-----------((((..-......-..))))----))......))))))))... ( -27.10, z-score = -3.36, R) >droAna3.scaffold_12943 484392 93 - 5039921 UCCGCUUUAC-ACAUUUUAUGUGCGCUCAAAA-GCCAAAUUCGUAGAGAUUU----GGCCGG-----------GUCGAA-AAACUU-GCGGAC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((.....-((((((((......)))))----))).((-----------(((...-......-...)))----))......))))))))... ( -28.60, z-score = -3.12, R) >dp4.chr4_group3 9607715 99 - 11692001 UUCGCUUUAC-ACGUUUUAUGUGCGCUCAAAA-GCCAAUUUUGUAGAGAUUU--GCUUUUGG------------UCCAA-AAACUUGCGGACCGGACCCAUUAUAAGCGCAUAAUC ..........-........((((((((.((((-((.((((((...)))))).--))))))((------------(((..-........)))))............))))))))... ( -26.20, z-score = -2.15, R) >droPer1.super_1 6722853 99 - 10282868 UUCGCUUUAC-ACGUUUUAUGUGCGCUCAAAA-GCCAAUUUUGUAGAGAUUU--GCUUUUGG------------UCCAA-AAACUUGCGGACCGGACCCAUUAUAAGCGCAUAAUC ..........-........((((((((.((((-((.((((((...)))))).--))))))((------------(((..-........)))))............))))))))... ( -26.20, z-score = -2.15, R) >droWil1.scaffold_180703 1876515 95 + 3946847 CUCCUUUUAC-ACAUUUUAUGUGCUCUCAAAA-GCCAAUUUUGUAGAGAUUU--GCUUUUGG------------UCCGA-AA-CUUGCGGCCC---UUCAUUAUAAGCGCAUAAUC ..........-.....(((((((((...((((-((.((((((...)))))).--))))))((------------.(((.-..-....))).))---.........))))))))).. ( -22.20, z-score = -2.54, R) >droVir3.scaffold_12963 10861988 104 - 20206255 CCCGCUUUAC-ACAUUUUAUGUGCGCUCAAAA-GCCAAUUUUGUAGAGAUUU-GGUAACGG---CCCAUGGCCACCGAGGAAACUU--GCGCC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((.....-(((((.(((....))).))-)))...((---((...))))..((((....)))--)....----........))))))))... ( -28.00, z-score = -1.76, R) >droMoj3.scaffold_6500 1939086 103 + 32352404 CCCGCUUUAC-ACAUUUUAUGUGCGCUCAAAA-GCCAAUUUUGUAGAGAUUUUGGUAACGGGAGCC-----CCGUUGAGGCAACUU--GCGCC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((.....-(((((..((.....))..)))))((((((...)-----)))))(.(((.....--..)))----.)......))))))))... ( -27.00, z-score = -1.22, R) >droGri2.scaffold_15252 12461734 110 + 17193109 CCCGCUUUAC-ACAUUUUAUGUGCGCUCAAAA-GCCAAUUUUGUAGAGAUUUUGGAAAAUGGCUCGAAUGGCCGUCGAGUAAACUCUGGCGCC----CCAUUAUAAGCGCAUAAUC ..........-........((((((((....(-((((.((((.((((...)))).)))))))))..(((((.(((((((....))).))))..----)))))...))))))))... ( -30.90, z-score = -1.93, R) >consensus CCCGCUUUAC_ACAUUUUAUGUGCGCUCAAAA_GCCAAUUUCGUAGAGAUUU____GGCCGG___________GUCCAA_AAACUU_GCGGAC____CCAUUAUAAGCGCAUAAUC ................((((((((((.......)).((((((...)))))).......................................................)))))))).. ( -8.99 = -8.91 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:59 2011