
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 489,994 – 490,058 |
| Length | 64 |
| Max. P | 0.690281 |

| Location | 489,994 – 490,058 |
|---|---|
| Length | 64 |
| Sequences | 4 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Shannon entropy | 0.36010 |
| G+C content | 0.40023 |
| Mean single sequence MFE | -15.76 |
| Consensus MFE | -9.71 |
| Energy contribution | -10.03 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 489994 64 + 2555491 GUAACGGUCAAACUUGCAUUUGUUUAAGCGGGCUAGUGUUCUUGUUCACAGACUGCUACAGUAU (((.(((((...(((((..........)))))...(((........))).))))).)))..... ( -14.90, z-score = -0.72, R) >droSec1.super_61 153577 64 + 189864 AUAACGGUCAGUCUUGUAUUUGUUUAAGCGGGUUACUGAUAUUUUUCCCAGACUGUUACAGUAU .((((((((.(.((((........)))))(((..............))).))))))))...... ( -14.34, z-score = -1.70, R) >droYak2.chrU 10697691 61 + 28119190 GUAGCGGUCGCACUU---UUGAUUUUAGUGGGCUACUGUUUUUGUUCACUGACUGCUACAGUAU (((((((((...(..---..).....(((((((..........))))))))))))))))..... ( -20.70, z-score = -3.47, R) >droEre2.scaffold_4784 375828 61 - 25762168 GUAGCGUACGCACUU---UUGAUUUUAGUGGACUACUGUUUUUGUUCACAGACUGCUACAGAAU ((((((....)....---.........((((((..........)))))).....)))))..... ( -13.10, z-score = -0.95, R) >consensus GUAACGGUCACACUU___UUGAUUUAAGCGGGCUACUGUUUUUGUUCACAGACUGCUACAGUAU (((((((((............((....))((((..........))))...)))))))))..... ( -9.71 = -10.03 + 0.31)

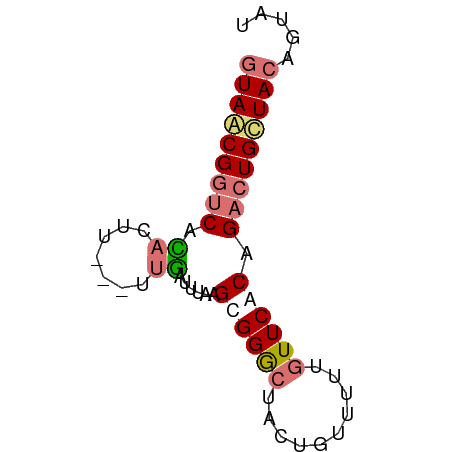
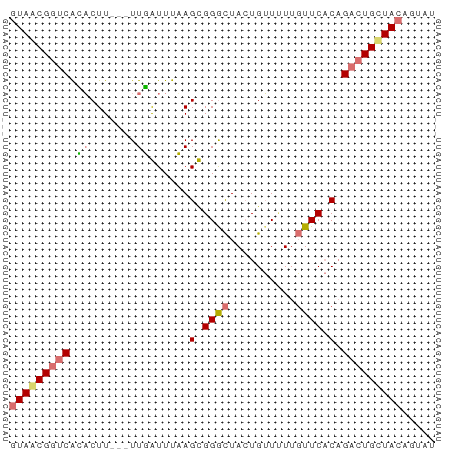
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:35 2011