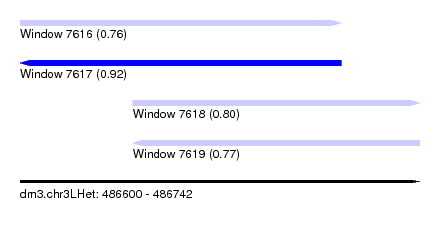
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 486,600 – 486,742 |
| Length | 142 |
| Max. P | 0.923076 |

| Location | 486,600 – 486,714 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.11 |
| Shannon entropy | 0.69432 |
| G+C content | 0.51941 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -8.65 |
| Energy contribution | -13.65 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.762760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 486600 114 + 2555491 UACCAGUAUUUAUUAACAUACGGCAGGCUUGCUCCUGCAAACCGUACUGUAGUGUGCCUACU-CUGGGCAAAAUGC-----AGCAGCAGCAUCGCCGUCUUGCUGGUGAGGACGGCAACG ((((((((...........(((((..(((.(((.(((((.....(((....)))(((((...-..)))))...)))-----)).))))))...)))))..))))))))....((....)) ( -40.12, z-score = -1.22, R) >droAna3.scaffold_13219 44901 114 - 86653 UACUUCACCUGUUUCGCAUACCUAAUGCUGGAGCCGUCGAGCCUGACAACGGGGGACCUG------GACAAGCUGCCUUUUAGCAGCAUUGUCACAGUCUUAUCCCUUGCCAAGCUGACU ..........(((((((((.....)))).))))).(((.(((.((.(((.(((((((.((------((((((((((......))))).))))).))))))...)))))).)).)))))). ( -40.60, z-score = -2.94, R) >droVir3.scaffold_12737 3614 120 - 48473 UAUUUCACCUCAGACGCAUACCUCACAUUCACUCCAUUCAGUCUGACAAUGGGUGGCCGACUAGCUGCCAAUCUGCCCUUGAGCAGCAUUGACACUGUCUUGUCCAUUGCAAGACUGACC .....................................(((((((..(((((((..(..(((.((.((.((((((((......)))).)))).))))))))..)))))))..))))))).. ( -33.90, z-score = -2.15, R) >droGri2.scaffold_15040 117899 120 + 162499 UAUUUCACCUCCGACACAUAUCUCAGGCUGCCGCCAUUUAGCCUAACCAAUGGGGGUCGACUACGCGACAAUCUGCCCUUAAGCAGCAUUGUCACGGUCUUGUCCAUUGCCAGACUGACG ....((((((((((........))((((((........)))))).......)))))).))...((.((((((((((......)))).)))))).))(((..(((........))).))). ( -36.60, z-score = -2.66, R) >consensus UACUUCACCUCAGACACAUACCUCAGGCUGGCGCCAUCAAGCCUGACAAUGGGGGGCCGACU_C__GACAAUCUGCCCUU_AGCAGCAUUGUCACAGUCUUGUCCAUUGCCAGACUGACG .........................(((((((....))))))).......................((((((((((......)))).)))))).(((((((((.....)))))))))... ( -8.65 = -13.65 + 5.00)

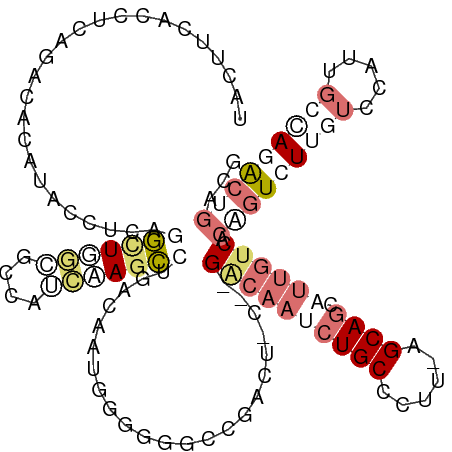
| Location | 486,600 – 486,714 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.11 |
| Shannon entropy | 0.69432 |
| G+C content | 0.51941 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -13.26 |
| Energy contribution | -15.57 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 486600 114 - 2555491 CGUUGCCGUCCUCACCAGCAAGACGGCGAUGCUGCUGCU-----GCAUUUUGCCCAG-AGUAGGCACACUACAGUACGGUUUGCAGGAGCAAGCCUGCCGUAUGUUAAUAAAUACUGGUA .(((((((((...........)))))))))((((((.((-----(((....(((...-.((((.....)))).....))).))))).))).))).((((((((........)))).)))) ( -40.90, z-score = -1.41, R) >droAna3.scaffold_13219 44901 114 + 86653 AGUCAGCUUGGCAAGGGAUAAGACUGUGACAAUGCUGCUAAAAGGCAGCUUGUC------CAGGUCCCCCGUUGUCAGGCUCGACGGCUCCAGCAUUAGGUAUGCGAAACAGGUGAAGUA .((((((((((((((((....((((..(((((.((((((....)))))))))))------..)))).))).)))))))))).))).......((((.....))))............... ( -46.00, z-score = -2.89, R) >droVir3.scaffold_12737 3614 120 + 48473 GGUCAGUCUUGCAAUGGACAAGACAGUGUCAAUGCUGCUCAAGGGCAGAUUGGCAGCUAGUCGGCCACCCAUUGUCAGACUGAAUGGAGUGAAUGUGAGGUAUGCGUCUGAGGUGAAAUA ..(((((((.(((((((....((((((((((((.(((((....)))))))))))).)).)))......))))))).))))))).........((.(.(((......))).).))...... ( -41.00, z-score = -1.36, R) >droGri2.scaffold_15040 117899 120 - 162499 CGUCAGUCUGGCAAUGGACAAGACCGUGACAAUGCUGCUUAAGGGCAGAUUGUCGCGUAGUCGACCCCCAUUGGUUAGGCUAAAUGGCGGCAGCCUGAGAUAUGUGUCGGAGGUGAAAUA (((((((((((((((((....((((((((((((.(((((....))))))))))))))..))).....))))).)))))))....)))))...((((..(((....)))..))))...... ( -46.50, z-score = -2.82, R) >consensus CGUCAGCCUGGCAAUGGACAAGACAGUGACAAUGCUGCU_AAGGGCAGAUUGUC__G_AGUAGGCCCCCCAUUGUCAGGCUAAAUGGAGCCAGCCUGAGGUAUGCGAAUGAGGUGAAAUA ....((((((((((.((((......))((((((.(((((....)))))))))))..............)).))))))))))....................................... (-13.26 = -15.57 + 2.31)

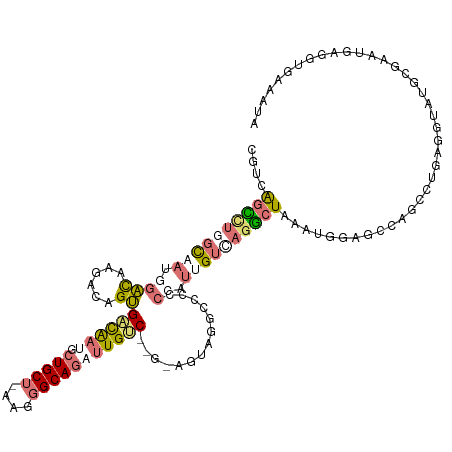
| Location | 486,640 – 486,742 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.48 |
| Shannon entropy | 0.63229 |
| G+C content | 0.55689 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -12.51 |
| Energy contribution | -16.32 |
| Covariance contribution | 3.81 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 486640 102 + 2555491 ACCGUACUGUAGUGUGCCUACU-CUGGGCAAAAUGC-----AGCAGCAGCAUCGCCGUCUUGC---UGGUGAG---GACGGCAACGCCGACUUCCCAUGCACCAACAACGGACA------ .((((.(((((...(((((...-..)))))...)))-----))..((((((.........)))---(((.(((---(.((((...)))).)))))))))).......))))...------ ( -36.40, z-score = -1.01, R) >droAna3.scaffold_13219 44941 114 - 86653 GCCUGACAACGGGGGACCUG------GACAAGCUGCCUUUUAGCAGCAUUGUCACAGUCUUAUCCCUUGCCAAGCUGACUCCCACACUGUCGCCCCACUGGCCAACAAUGCGCAAGCACU ((.((.(((.(((((((.((------((((((((((......))))).))))).))))))...)))))).)).)).(((.........)))(((.....)))......(((....))).. ( -38.80, z-score = -2.19, R) >droVir3.scaffold_12737 3654 114 - 48473 GUCUGACAAUGGGUGGCCGACUAGCUGCCAAUCUGCCCUUGAGCAGCAUUGACACUGUCUUGUCCAUUGCAAGACUGACCCCAACACUUUCACCCCACUCAUACACAGUGCGCA------ (((((...((((((((.......((((((((.......))).))))).........(((((((.....)))))))...................))))))))...))).))...------ ( -31.50, z-score = -1.09, R) >droGri2.scaffold_15040 117939 114 + 162499 GCCUAACCAAUGGGGGUCGACUACGCGACAAUCUGCCCUUAAGCAGCAUUGUCACGGUCUUGUCCAUUGCCAGACUGACGCCAACACUGAGACCCCACUCAUACACAACUCGCA------ ............((((((......((((((((((((......)))).)))))).((((((.((.....)).))))))..))((....)).))))))..................------ ( -33.90, z-score = -2.60, R) >consensus GCCUGACAAUGGGGGGCCGACU_C__GACAAUCUGCCCUU_AGCAGCAUUGUCACAGUCUUGUCCAUUGCCAGACUGACGCCAACACUGACACCCCACUCACAAACAACGCGCA______ ...........((((...........((((((((((......)))).)))))).(((((((((.....)))))))))...............))))........................ (-12.51 = -16.32 + 3.81)

| Location | 486,640 – 486,742 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.48 |
| Shannon entropy | 0.63229 |
| G+C content | 0.55689 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -13.04 |
| Energy contribution | -15.47 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 486640 102 - 2555491 ------UGUCCGUUGUUGGUGCAUGGGAAGUCGGCGUUGCCGUC---CUCACCA---GCAAGACGGCGAUGCUGCUGCU-----GCAUUUUGCCCAG-AGUAGGCACACUACAGUACGGU ------...((((..((((.(((..(..((((((((((((((((---.......---....)))))))))))))..)))-----...)..)))))))-.((((.....))))...)))). ( -39.20, z-score = -0.76, R) >droAna3.scaffold_13219 44941 114 + 86653 AGUGCUUGCGCAUUGUUGGCCAGUGGGGCGACAGUGUGGGAGUCAGCUUGGCAAGGGAUAAGACUGUGACAAUGCUGCUAAAAGGCAGCUUGUC------CAGGUCCCCCGUUGUCAGGC ...((((.((((((((((.((....)).)))))))))).))))..((((((((((((....((((..(((((.((((((....)))))))))))------..)))).))).))))))))) ( -58.50, z-score = -4.95, R) >droVir3.scaffold_12737 3654 114 + 48473 ------UGCGCACUGUGUAUGAGUGGGGUGAAAGUGUUGGGGUCAGUCUUGCAAUGGACAAGACAGUGUCAAUGCUGCUCAAGGGCAGAUUGGCAGCUAGUCGGCCACCCAUUGUCAGAC ------..(.((((.......)))).).(((.((((..((((((.((((......))))..((((((((((((.(((((....)))))))))))).)).))))))).)))))).)))... ( -40.40, z-score = -0.83, R) >droGri2.scaffold_15040 117939 114 - 162499 ------UGCGAGUUGUGUAUGAGUGGGGUCUCAGUGUUGGCGUCAGUCUGGCAAUGGACAAGACCGUGACAAUGCUGCUUAAGGGCAGAUUGUCGCGUAGUCGACCCCCAUUGGUUAGGC ------.....(((....((.((((((((((((.(((..((....).)..))).)))....((((((((((((.(((((....))))))))))))))..))))).))))))).))..))) ( -42.70, z-score = -1.83, R) >consensus ______UGCGCAUUGUGGAUGAGUGGGGUGACAGUGUUGGCGUCAGUCUGGCAAUGGACAAGACAGUGACAAUGCUGCU_AAGGGCAGAUUGUC__G_AGUAGGCCCCCCAUUGUCAGGC .........((((((((.((.......))))))))))........(((((((((.((((......))((((((.(((((....)))))))))))..............)).))))))))) (-13.04 = -15.47 + 2.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:33 2011