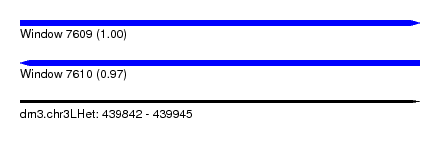
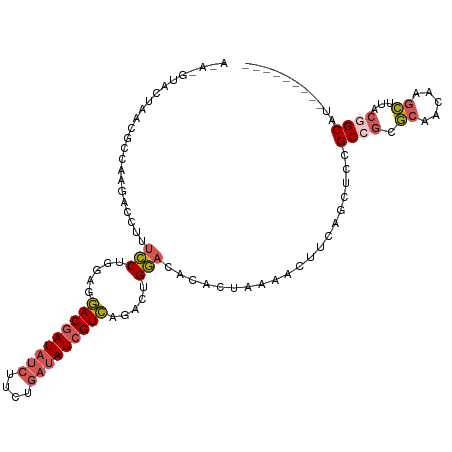
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 439,842 – 439,945 |
| Length | 103 |
| Max. P | 0.997904 |

| Location | 439,842 – 439,945 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.91 |
| Shannon entropy | 0.52170 |
| G+C content | 0.49593 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 439842 103 + 2555491 ACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUAAAACUUCAGCUCCGC-GCGCAACAAGCUUACGGCAU--------- ............(((............(((((((((((((....)))))))))...(((((...........)))))))))..-((.......))....)))..--------- ( -28.60, z-score = -2.60, R) >droAna3.scaffold_13250 842529 109 - 3535662 AUAUGUGGCAACCCUAAG-CGUUUACCAUAAUACAAUCGAACUCGAUAUCGUAUGAGAGUAU-CGAUAUCGCUUUUUCUGCGCCUCACGACA--UCGACAGCCUCAUCCGCAG ..(((.(((........(-..((......))..)..((((..((((((((((((....))).-)))))))((.......)).......))..--))))..))).)))...... ( -23.20, z-score = -0.88, R) >droSim1.chr2h_random 683859 104 + 3178526 ACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUUAAACUUCAGCUCCGCCGCGCAACAAGCUUACGGCAU--------- ..((((.((.......)))))).....(((((((((((((....)))))))))...(((((...........)))))))))((((.((.....))...))))..--------- ( -31.80, z-score = -3.55, R) >droSec1.super_0 589422 98 + 21120651 ------ACAAGUACCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGGAACUUCAGCUCUGCCGCGCAACAAGCUAACGGCAU--------- ------...................(((((((((((((((....)))))))))...(..(.....)..)..))))))...(((((.((.....))...))))).--------- ( -29.70, z-score = -2.77, R) >droYak2.chrU 11905997 97 - 28119190 ------ACUAACGCCAAGACCUUUCCUGGAUGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACUCU-GGACUUCAACCCCGCCGCGCAACAAGCUUACGGCAU--------- ------......((((...((......)).((((((((((....))))))))))...))))......-((.........))((((.((.....))...))))..--------- ( -29.40, z-score = -3.75, R) >consensus A_A_GUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUAAAACUUCAGCUCCGCCGCGCAACAAGCUUACGGCAU_________ .......................(((.....(((((((((....))))))))).....)))....................((((.((.....))...))))........... (-15.64 = -16.48 + 0.84)

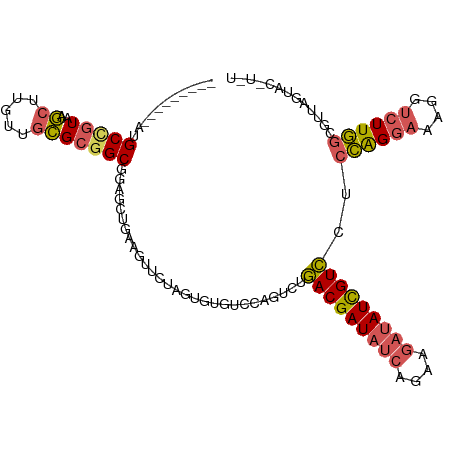
| Location | 439,842 – 439,945 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.91 |
| Shannon entropy | 0.52170 |
| G+C content | 0.49593 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -19.74 |
| Energy contribution | -20.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 439842 103 - 2555491 ---------AUGCCGUAAGCUUGUUGCGC-GCGGAGCUGAAGUUUUAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGU ---------((((((..(.(((....(((-(((((((....))))).)))))(((.....(((((((((....))))))))).....)))))).)..)))))).......... ( -31.30, z-score = -1.28, R) >droAna3.scaffold_13250 842529 109 + 3535662 CUGCGGAUGAGGCUGUCGA--UGUCGUGAGGCGCAGAAAAAGCGAUAUCG-AUACUCUCAUACGAUAUCGAGUUCGAUUGUAUUAUGGUAAACG-CUUAGGGUUGCCACAUAU ...((.(((((..((((((--((((((..............)))))))))-)))..))))).))..((((....)))).......((((((.(.-....)..))))))..... ( -32.24, z-score = -1.03, R) >droSim1.chr2h_random 683859 104 - 3178526 ---------AUGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGUUUAAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGU ---------..(((((..((.....)))))))((.(((((.(((.(((....(((.....(((((((((....))))))))).....))).....)))))).))))).))... ( -35.10, z-score = -2.19, R) >droSec1.super_0 589422 98 - 21120651 ---------AUGCCGUUAGCUUGUUGCGCGGCAGAGCUGAAGUUCCAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGUACUUGU------ ---------.((((((..((.....))))))))((((....)))).((((..........(((((((((....)))))))))..((((((....))))))))))...------ ( -32.20, z-score = -1.96, R) >droYak2.chrU 11905997 97 + 28119190 ---------AUGCCGUAAGCUUGUUGCGCGGCGGGGUUGAAGUCC-AGAGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCAUCCAGGAAAGGUCUUGGCGUUAGU------ ---------.((((((..((.....))))))))...((((.(((.-(((.(..((.(..((((((((((....))))))))))..).))...).))).))).)))).------ ( -34.10, z-score = -2.03, R) >consensus _________AUGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGUUCUAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUAC_U_U ...........(((((..((.....)))))))............................(((((((((....)))))))))..((((((....))))))............. (-19.74 = -20.14 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:26 2011