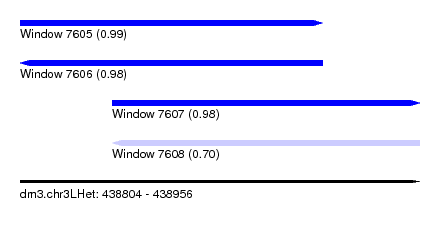
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 438,804 – 438,956 |
| Length | 152 |
| Max. P | 0.994077 |

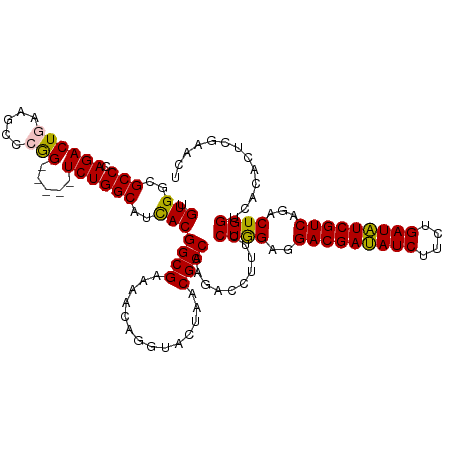
| Location | 438,804 – 438,919 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Shannon entropy | 0.18306 |
| G+C content | 0.53630 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -32.16 |
| Energy contribution | -32.32 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 438804 115 + 2555491 UGUGGCGCCCAGACUGAAGCCCGG-----UCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUAAAACU .(((..(((.((((((.....)))-----))))))..)))((((..............))))........(((.(...(((((((((....)))))))))...).)))............ ( -40.44, z-score = -3.32, R) >droSim1.chr2h_random 682888 115 + 3178526 UGUGGCGCCCAGACUGAAGCCCGG-----UCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACU (((((((((.((((((.....)))-----)))))).((.(((.(((...((((.((.......)))))))))))))).(((((((((....)))))))))......))))))........ ( -43.80, z-score = -3.26, R) >droSec1.super_163 39661 115 + 51592 UGUGGCGCCCAGACUGAAGCCCGG-----UCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGGCUCACUGGAACU ........((((..(((.((((((-----((((((.....((((..............))))....((((.....))))..((((((....))))))))))))))))))))))))).... ( -47.04, z-score = -3.84, R) >droYak2.chrU 21900759 119 + 28119190 UGUGGCGCCCAGACUAAAGCCCAGCCCAGUCUGGCAAUACGGCGUAAACAAGUACUAACGCCAAGACCUGUCCUAGAGGACGACAUCUUCUGAUGUCGUCAGACUGGUCACUCUCGACA- ((.(((............)))))(.((((((((((.....(((((............))))).......((((....))))((((((....)))))))))))))))).)..........- ( -44.30, z-score = -3.48, R) >droEre2.scaffold_4929 4656536 114 - 26641161 UGUGGCGCCCAGACUAAAGCCCAG-----UCUGGCAACACGGCGUAAGCAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGUCACUCUCGACU- .(((..(((.(((((.......))-----))))))..)))(((((.((......)).)))))..((((..((......(((((((((....))))))))).))..))))..........- ( -42.90, z-score = -3.60, R) >consensus UGUGGCGCCCAGACUGAAGCCCGG_____UCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGUCACACUCGAACU .(((((..((((......(((.((......))))).....((((..............))))..........))))..(((((((((....)))))))))......)))))......... (-32.16 = -32.32 + 0.16)

| Location | 438,804 – 438,919 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.39 |
| Shannon entropy | 0.18306 |
| G+C content | 0.53630 |
| Mean single sequence MFE | -46.15 |
| Consensus MFE | -34.68 |
| Energy contribution | -34.48 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.982033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 438804 115 - 2555491 AGUUUUAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGA-----CCGGGCUUCAGUCUGGGCGCCACA .......((((((((((.(((((((((((....)))))))(((....)))..(((((.(((((((................))))))))))-----)).....)))).))))))).))). ( -45.49, z-score = -3.64, R) >droSim1.chr2h_random 682888 115 - 3178526 AGUUCCAGUGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGA-----CCGGGCUUCAGUCUGGGCGCCACA .......(((((.((((.(((((((((((....)))))))(((....)))..(((((.(((((((................))))))))))-----)).....)))).)))).)).))). ( -43.59, z-score = -2.29, R) >droSec1.super_163 39661 115 - 51592 AGUUCCAGUGAGCCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGA-----CCGGGCUUCAGUCUGGGCGCCACA .......(((.((((((.(((((((((((....)))))))(((....)))..(((((.(((((((................))))))))))-----)).....)))).))))))..))). ( -47.49, z-score = -3.73, R) >droYak2.chrU 21900759 119 - 28119190 -UGUCGAGAGUGACCAGUCUGACGACAUCAGAAGAUGUCGUCCUCUAGGACAGGUCUUGGCGUUAGUACUUGUUUACGCCGUAUUGCCAGACUGGGCUGGGCUUUAGUCUGGGCGCCACA -..........((((.(((((((((((((....))))))))).....)))).)))).((((((((((((...........))))))(((((((((((...)).))))))))))))))).. ( -47.30, z-score = -2.17, R) >droEre2.scaffold_4929 4656536 114 + 26641161 -AGUCGAGAGUGACCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGCUUACGCCGUGUUGCCAGA-----CUGGGCUUUAGUCUGGGCGCCACA -.(((.(((.(((((((((((((((((((....)))))))))..........(((..((((((.(((....))).))))))....))))))-----))))...))).))).)))...... ( -46.90, z-score = -3.28, R) >consensus AGUUCCAGUGUGACCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGA_____CCGGGCUUCAGUCUGGGCGCCACA ...........((((..((((((((((((....))))))))).....)))..))))..((((..((......))..))))(((.((((.........(((((....))))))))).))). (-34.68 = -34.48 + -0.20)

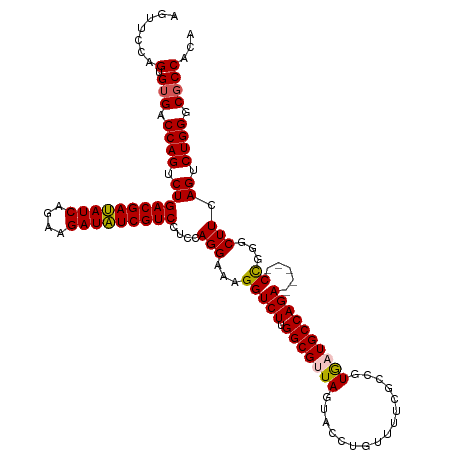
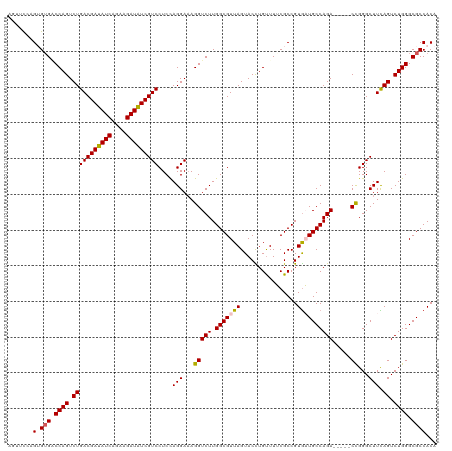
| Location | 438,839 – 438,956 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.15 |
| Shannon entropy | 0.22664 |
| G+C content | 0.48983 |
| Mean single sequence MFE | -34.37 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 438839 117 + 2555491 GGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUAAAACUUCAGCUCCGC-GCGCAACAAGCUUACGGCA--UAAAACAA .(((.....((((.((.......)))))).....(((((((((((((....)))))))))...(((((...........))))))))).)-)).......((.....)).--........ ( -29.80, z-score = -2.28, R) >droSim1.chr2h_random 682923 119 + 3178526 GGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACUUCAACGCCGC-GCGCAACAAGCUUACUUUAAUCUAAACAA ((((.....((((.((.......))))))....((((((((((((((....)))))))))...(..(.....)..)..))))).))))..-((.......)).................. ( -31.10, z-score = -2.41, R) >droSec1.super_163 39696 118 + 51592 GGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGGCUCACUGGAACUUCAGCUCCGCCGCGCAACAAGCUUACGGCA--UAAAACAA ((((..............))))..........(((((((((((((((....)))))))))...(..(.....)..)..))))))....((((.((.....))...)))).--........ ( -35.74, z-score = -2.53, R) >droYak2.chrU 21900799 118 + 28119190 GGCGUAAACAAGUACUAACGCCAAGACCUGUCCUAGAGGACGACAUCUUCUGAUGUCGUCAGACUGGUCACUCUCGA-CAUUAGCUGCGCCGCGAAACAAGCUUACGGCA-UCAAAUUAA (((((............)))))..((((.(((......(((((((((....))))))))).))).))))........-..........((((..(.......)..)))).-......... ( -37.40, z-score = -2.91, R) >droEre2.scaffold_4929 4656571 118 - 26641161 GGCGUAAGCAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGUCACUCUCGA-CUUCAGCCUUGCCGCGCAACAAGCUUACGGCA-UCAAAUUAA (((((.((......)).)))))..........(((((((((((((((....))))))))).((....))........-))))))...(((((.((.....))...)))))-......... ( -37.80, z-score = -3.23, R) >consensus GGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGUCACACUCGAACUUCAGCUCCGCCGCGCAACAAGCUUACGGCA_UCAAAACAA ((((..............))))..........(((((((((((((((....)))))))))...((((.....))))..))))))....((((.((.....))...))))........... (-26.86 = -27.78 + 0.92)

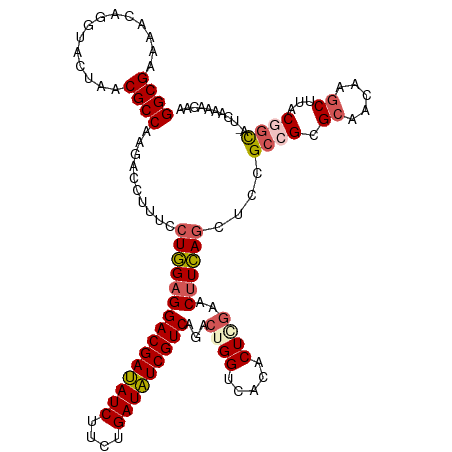
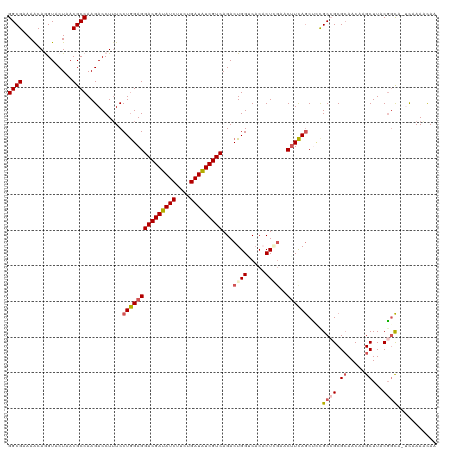
| Location | 438,839 – 438,956 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Shannon entropy | 0.22664 |
| G+C content | 0.48983 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -27.14 |
| Energy contribution | -28.22 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 438839 117 - 2555491 UUGUUUUA--UGCCGUAAGCUUGUUGCGC-GCGGAGCUGAAGUUUUAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCC ........--..((((..((.....))..-)))).((.((((...(((.(((........(((((((((....)))))))))(.((((((....)))))).)....)))))).)))))). ( -32.10, z-score = -0.83, R) >droSim1.chr2h_random 682923 119 - 3178526 UUGUUUAGAUUAAAGUAAGCUUGUUGCGC-GCGGCGUUGAAGUUCCAGUGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCC ..............(.((((..((.((((-((.((((((......)))))).))......(((((((((....)))))))))...(((((....)))))))))....))..)))).)... ( -30.60, z-score = -0.10, R) >droSec1.super_163 39696 118 - 51592 UUGUUUUA--UGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGUUCCAGUGAGCCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCC ........--.(((((..((.....)))))))(((((....))))).(((((..(((...(((((((((....)))))))))(.((((((....)))))).).......)))..))))). ( -42.60, z-score = -3.15, R) >droYak2.chrU 21900799 118 - 28119190 UUAAUUUGA-UGCCGUAAGCUUGUUUCGCGGCGCAGCUAAUG-UCGAGAGUGACCAGUCUGACGACAUCAGAAGAUGUCGUCCUCUAGGACAGGUCUUGGCGUUAGUACUUGUUUACGCC ......((.-(((((((((....))).))))))))(((((((-((((((.(..((((...(((((((((....)))))))))..)).))...).)))))))))))))............. ( -43.90, z-score = -3.18, R) >droEre2.scaffold_4929 4656571 118 + 26641161 UUAAUUUGA-UGCCGUAAGCUUGUUGCGCGGCAAGGCUGAAG-UCGAGAGUGACCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGCUUACGCC .........-...(((((((..((...(((.(((((((....-((..(((.((....)).(((((((((....))))))))))))..))...))))))).)))....))..))))))).. ( -42.10, z-score = -2.79, R) >consensus UUGUUUUGA_UGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGUUCCAGUGUGACCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCC ..........((((((..((.....))))))))..............((((((.(((...(((((((((....)))))))))(.((((((....)))))).).......))).)))))). (-27.14 = -28.22 + 1.08)

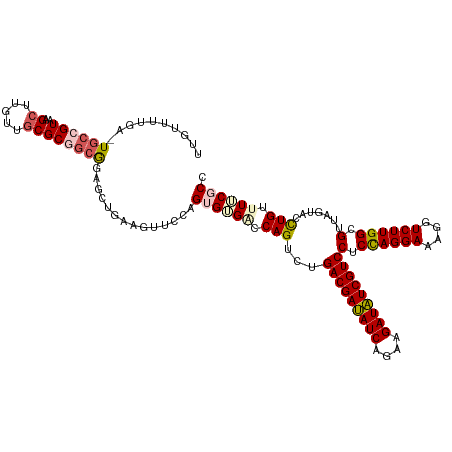
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:24 2011