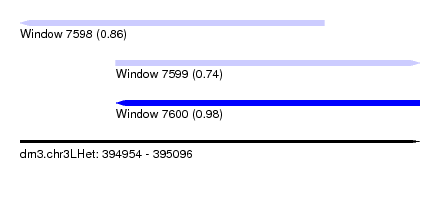
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 394,954 – 395,096 |
| Length | 142 |
| Max. P | 0.976010 |

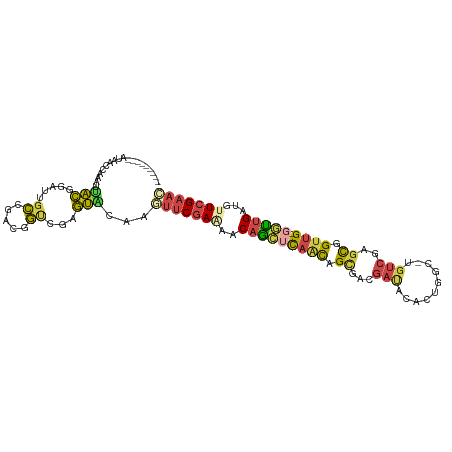
| Location | 394,954 – 395,062 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 59.42 |
| Shannon entropy | 0.75493 |
| G+C content | 0.50607 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -10.60 |
| Energy contribution | -10.42 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
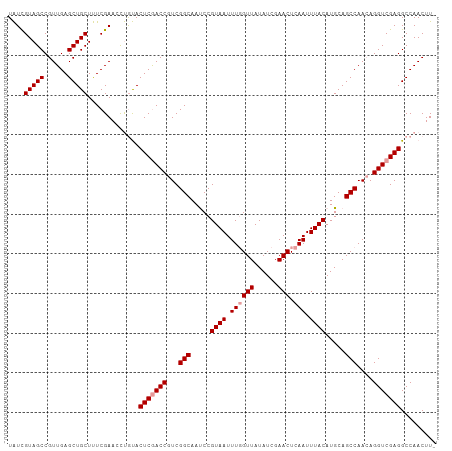
>dm3.chr3LHet 394954 108 - 2555491 UGAGUUCGAUAUAACGAAAUUACGGGUUGCCGACGGUCGAGUACAGGUUCGAAAGCAGCUCAACGGCUACAAUACGCUGAC--GUCGAGCGGUUG-GGUUGAUGUUCGAAC ((..((((((.(((.....)))(((....)))...))))))..)).(((((((..(((((((((.(((.....(((....)--))..))).))))-)))))...))))))) ( -34.40, z-score = -1.21, R) >droWil1.scaffold_181099 48770 100 - 85210 ----------AUGAAAUAUUUGCAAAGAGGCAACACGUUAGCAGAAGAGCAACAACAUAGUUUCAGAGACGAAAAGGAUGG-UGCCAGGUGGCCGCAGGAGCAGAUCGGAG ----------.......((((((.....((((....(((.((......)))))..(((..((((......))))...))).-))))..((....))....))))))..... ( -18.70, z-score = -0.14, R) >droSim1.chr2h_random 2617553 108 - 3178526 UGAGUUCGAUAUAACCAAAUUACGGAUUGCCGACGGUUGAGUGUGGGUUCGAGAGCAGCUCAACGGCUACGACACACUGGC--GUCGAGCGGUUG-UGUUGAUGUUCGAAC .((..(((((((((((......(((....)))...(((((((....(((....))).))))))).(((.((((.(....).--))))))))))))-))))))...)).... ( -37.20, z-score = -1.57, R) >droSec1.super_65 4126 110 + 171001 UGAGUUCGAUAUAACCAAAUUACGGAUUGCCGACGGUCGAGUACAAUUUCGAAAGCAGCUCAACGGCUACGAUACACUGGCGUGUCGAGCGGUUG-GGUUGAUGUUCGAAC ((..((((((.(((.....)))(((....)))...))))))..))..((((((..(((((((((.(((.((((((......))))))))).))))-)))))...)))))). ( -40.30, z-score = -3.27, R) >droYak2.chr3h_random 357177 97 + 911842 ------------AACUCGGACAAGUUGCGCUAGUUGUCGAUUGC-ACUUCGAAGACAGUUCAGCAGUGACGAUACACUGGCGUGUCUAGUUGUUG-GACUGAUGUUCGAAG ------------......(((((.(......).)))))......-.(((((((.((((((((((((..(.(((((......))))))..))))))-))))).).))))))) ( -32.90, z-score = -1.46, R) >droEre2.scaffold_4929 2234936 94 + 26641161 -------------ACUCAA--ACGGCUUGCCGACGGCCAAGUUCGAGUUCGAGUACAGCUCAAUAGCGACGAUCCACUGGCGUGUCUAGCUGUUG-GGCUGAUG-UCGAAC -------------......--.(((((((((...)).)))).))).((((((...((((((((((((((((..((...))..))))..)))))))-)))))...-)))))) ( -35.50, z-score = -2.00, R) >consensus __________AUAACCAAAUUACGGAUUGCCGACGGUCGAGUACAAGUUCGAAAACAGCUCAACAGCGACGAUACACUGGC_UGUCGAGCGGUUG_GGUUGAUGUUCGAAC ....................(((.....((.....))...)))...(((((((..(((((((((.((.....................)).)))).)))))...))))))) (-10.60 = -10.42 + -0.18)

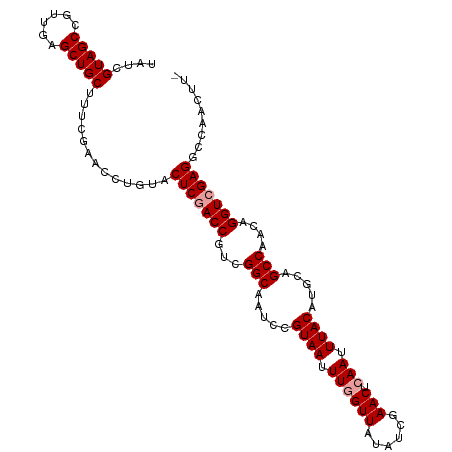
| Location | 394,988 – 395,096 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Shannon entropy | 0.11671 |
| G+C content | 0.47066 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -25.13 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 394988 108 + 2555491 UAUUGUAGCCGUUGAGCUGCUUUCGAACCUGUACUCGACCGUCGGCAACCCGUAAUUUCGUUAUAUCGAACUCAAUUUACAUGCAGCCAACAGGUCGAGGCCAACUUA ....(((((......))))).........((..(((((((((.(((.....((((.((((......))))......)))).....))).)).)))))))..))..... ( -28.60, z-score = -1.73, R) >droSim1.chr2h_random 2617587 107 + 3178526 UGUCGUAGCCGUUGAGCUGCUCUCGAACCCACACUCAACCGUCGGCAAUCCGUAAUUUGGUUAUAUCGAACUCAAUUUACAUGCAGCCAACAGGUCGAGGCCAACUU- .(((...(((((((.(((((.......((.((........)).))......((((.((((((......))).))).))))..))))))))).)))...)))......- ( -27.10, z-score = -1.15, R) >droSec1.super_65 4162 108 - 171001 UAUCGUAGCCGUUGAGCUGCUUUCGAAAUUGUACUCGACCGUCGGCAAUCCGUAAUUUGGUUAUAUCGAACUCAAUUUACAUGCAGCCAAAAGGUCGAGGCCAACUUU ....(((((......)))))........(((..(((((((...(((.....((((.((((((......))).))).)))).....)))....)))))))..))).... ( -31.60, z-score = -2.50, R) >consensus UAUCGUAGCCGUUGAGCUGCUUUCGAACCUGUACUCGACCGUCGGCAAUCCGUAAUUUGGUUAUAUCGAACUCAAUUUACAUGCAGCCAACAGGUCGAGGCCAACUU_ ....(((((......))))).............(((((((...(((.....((((.((((((......))).))).)))).....)))....)))))))......... (-25.13 = -25.80 + 0.67)

| Location | 394,988 – 395,096 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Shannon entropy | 0.11671 |
| G+C content | 0.47066 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -31.11 |
| Energy contribution | -31.67 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
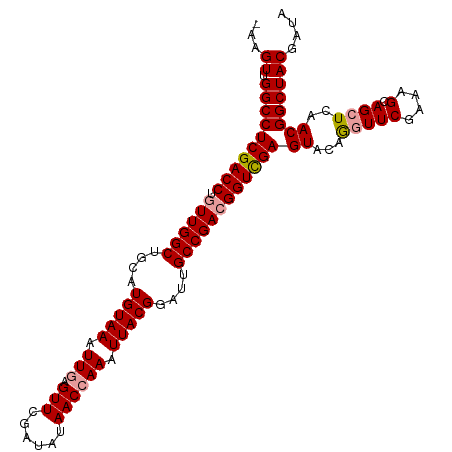
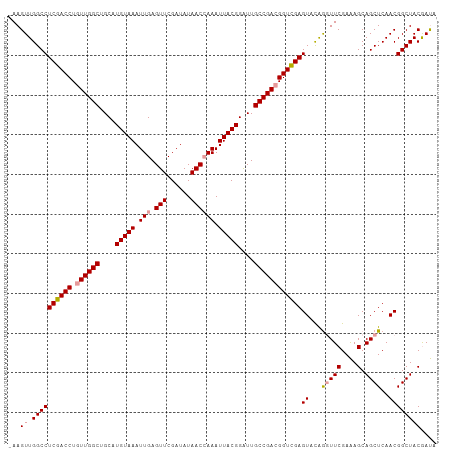
>dm3.chr3LHet 394988 108 - 2555491 UAAGUUGGCCUCGACCUGUUGGCUGCAUGUAAAUUGAGUUCGAUAUAACGAAAUUACGGGUUGCCGACGGUCGAGUACAGGUUCGAAAGCAGCUCAACGGCUACAAUA ...((.((((((((((.((((((.((.(((((......((((......)))).))))).)).))))))))))))((...(((((....).))))..)))))))).... ( -36.60, z-score = -2.61, R) >droSim1.chr2h_random 2617587 107 - 3178526 -AAGUUGGCCUCGACCUGUUGGCUGCAUGUAAAUUGAGUUCGAUAUAACCAAAUUACGGAUUGCCGACGGUUGAGUGUGGGUUCGAGAGCAGCUCAACGGCUACGACA -..(((((((((((((.((((((....(((((.(((.(((......)))))).)))))....))))))))))))((.(((((((....).)))))))))))..)))). ( -39.10, z-score = -2.86, R) >droSec1.super_65 4162 108 + 171001 AAAGUUGGCCUCGACCUUUUGGCUGCAUGUAAAUUGAGUUCGAUAUAACCAAAUUACGGAUUGCCGACGGUCGAGUACAAUUUCGAAAGCAGCUCAACGGCUACGAUA .((((((..(((((((..(((((....(((((.(((.(((......)))))).)))))....))))).)))))))..)))))).....(.((((....)))).).... ( -33.80, z-score = -2.76, R) >consensus _AAGUUGGCCUCGACCUGUUGGCUGCAUGUAAAUUGAGUUCGAUAUAACCAAAUUACGGAUUGCCGACGGUCGAGUACAGGUUCGAAAGCAGCUCAACGGCUACGAUA ...((.((((((((((.((((((....(((((.(((.(((......)))))).)))))....))))))))))))((...(((((....).))))..)))))))).... (-31.11 = -31.67 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:18 2011