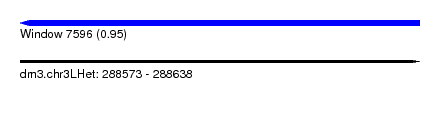
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 288,573 – 288,638 |
| Length | 65 |
| Max. P | 0.951871 |

| Location | 288,573 – 288,638 |
|---|---|
| Length | 65 |
| Sequences | 5 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 55.16 |
| Shannon entropy | 0.83840 |
| G+C content | 0.54471 |
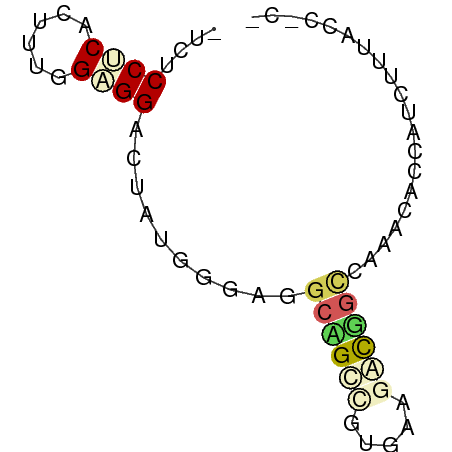
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -6.62 |
| Energy contribution | -7.30 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.70 |
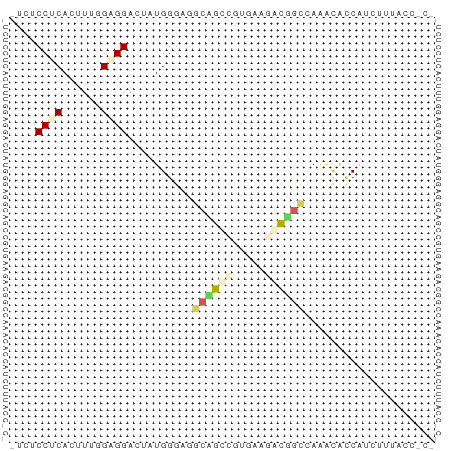
| Mean z-score | -1.55 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
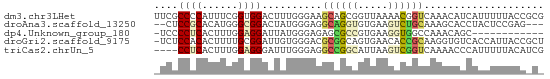
>dm3.chr3LHet 288573 65 - 2555491 UUCGCCCCAUUUCGGUGGACUUUGGGAAGCAGCGGUUAAAACGGUCAAACAUCAUUUUUACCGCG ...((((((..((....))...))))..)).(((((.((((.(((.....))).)))).))))). ( -17.10, z-score = -1.06, R) >droAna3.scaffold_13250 2026583 60 - 3535662 --CUCCGCACAUGGGCGGACUAUGGGAGGCAGGUGUGAAGUCUGCAAAGCACCUACUCCGAG--- --.(((((......))))).....((((..((((((...(....)...)))))).))))...--- ( -22.90, z-score = -1.79, R) >dp4.Unknown_group_180 23541 52 + 35195 -UCCCCUCACUUUGGAGGAUUAUGGGAGAGCGCCGUGAAGGUGGCCAAACAGC------------ -...((((......))))......((....((((.....)))).)).......------------ ( -11.80, z-score = 0.89, R) >droGri2.scaffold_9175 4771 64 + 5175 -UCUCCACACUUUUGCGGAUUGUGGGACGCGGCAGUGAACACCGCAAGGUGUCACCAUUACCGCU -..((((((.((.....)).))))))..((((.((((.(((((....)))))...)))).)))). ( -25.80, z-score = -3.03, R) >triCas2.chrUn_5 439372 61 - 975455 ----CCUCACUUUGGAGGGAUUUGGGAGGCCGGCAUUAAGUCGGUCAAAACCCAUUUUUACAUCG ----((((......))))((..((((.(((((((.....)))))))....))))..))....... ( -20.80, z-score = -2.77, R) >consensus _UCUCCUCACUUUGGAGGACUAUGGGAGGCAGCCGUGAAGACGGCCAAACACCAUCUUUACC_C_ ....((((......))))..........((((((.....)))))).................... ( -6.62 = -7.30 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:15 2011