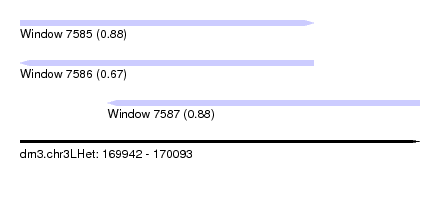
| Sequence ID | dm3.chr3LHet |
|---|---|
| Location | 169,942 – 170,093 |
| Length | 151 |
| Max. P | 0.882119 |

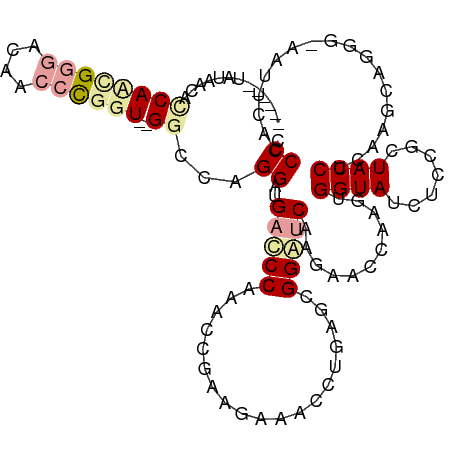
| Location | 169,942 – 170,053 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 58.55 |
| Shannon entropy | 0.58280 |
| G+C content | 0.56329 |
| Mean single sequence MFE | -47.77 |
| Consensus MFE | -8.67 |
| Energy contribution | -7.80 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.53 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.18 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.880043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 169942 111 + 2555491 ------UAAAAACCCAUUGCCGCAACUUGGUUAGCCCGGGAUGACCCUAACCGUAAGGGGCUGAGCGGGGCCAGAACGAAGUGGUAUCUUCGUUACCUUUAGCAGGCCAAGUCACCC- ------............(((((..((((.((((((((((....)))....(....)))))))).)))).....(((((((......))))))).......)).)))..........- ( -36.50, z-score = -0.50, R) >droVir3.scaffold_12823 181988 109 - 2474545 ------UAUAACGUCACCGGGACAACCCGGU--GGCAAGGGUGAUCCAAACAGAAGAAACCUGAGCGGAUCGCGCACCAAAUGGUAUGUGCGCUACCUCAAAAAGGG-AUUUCAUCCC ------......(((((((((....))))))--)))..(((((((((...(((.......)))...)))))((((((..........)))))).))))......(((-((...))))) ( -43.50, z-score = -4.24, R) >droGri2.scaffold_4812 5858 115 + 6386 AGAGCUCCUGCCACCAAGGGGUCAACCCGGU--GGGCUGGAUGACCCGAACCGGAGAAAUCUCUGCGGUUCAAGGGUCAAGUGGUACCUCCGGUACCUCCAGCAGGG-ACUCAACCCC .(((..(((.(((((..(((.....))))))--))((((((((((((((((((.(((....))).))))))..))))))...((((((...))))))))))))))).-.)))...... ( -63.30, z-score = -5.01, R) >consensus ______UAUAACACCAACGGGACAACCCGGU__GGCCAGGAUGACCCAAACCGAAGAAACCUGAGCGGAUCAAGAACCAAGUGGUAUCUCCGCUACCUCAAGCAGGG_AAUUCACCCC ............(((.(((((......((((......(((.....))).))))......)))))...)))............((((.......))))..................... ( -8.67 = -7.80 + -0.87)

| Location | 169,942 – 170,053 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 58.55 |
| Shannon entropy | 0.58280 |
| G+C content | 0.56329 |
| Mean single sequence MFE | -47.37 |
| Consensus MFE | -11.45 |
| Energy contribution | -8.69 |
| Covariance contribution | -2.76 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3LHet 169942 111 - 2555491 -GGGUGACUUGGCCUGCUAAAGGUAACGAAGAUACCACUUCGUUCUGGCCCCGCUCAGCCCCUUACGGUUAGGGUCAUCCCGGGCUAACCAAGUUGCGGCAAUGGGUUUUUA------ -..(..(((((((((.....))).(((((((......))))))).((((((.....((((......)))).(((....))))))))).))))))..)...............------ ( -37.30, z-score = 0.01, R) >droVir3.scaffold_12823 181988 109 + 2474545 GGGAUGAAAU-CCCUUUUUGAGGUAGCGCACAUACCAUUUGGUGCGCGAUCCGCUCAGGUUUCUUCUGUUUGGAUCACCCUUGCC--ACCGGGUUGUCCCGGUGACGUUAUA------ (((((...))-))).....((((..((((((..........))))))((((((..((((.....))))..))))))..))))..(--((((((....)))))))........------ ( -45.70, z-score = -4.15, R) >droGri2.scaffold_4812 5858 115 - 6386 GGGGUUGAGU-CCCUGCUGGAGGUACCGGAGGUACCACUUGACCCUUGAACCGCAGAGAUUUCUCCGGUUCGGGUCAUCCAGCCC--ACCGGGUUGACCCCUUGGUGGCAGGAGCUCU ......(((.-.(((((((((((((((...))))))...((((((..((((((..(((....)))))))))))))))))))))((--((((((....)))...))))).)))..))). ( -59.10, z-score = -3.33, R) >consensus GGGGUGAAAU_CCCUGCUAGAGGUAACGAAGAUACCACUUGGUCCUCGAACCGCUCAGAUUUCUACGGUUAGGGUCAUCCAGGCC__ACCGGGUUGACCCAAUGGCGUUAUA______ .(((........(((.....)))...(((.(((........))).))).)))......(((....(..(((((...............)))))..).......)))............ (-11.45 = -8.69 + -2.76)

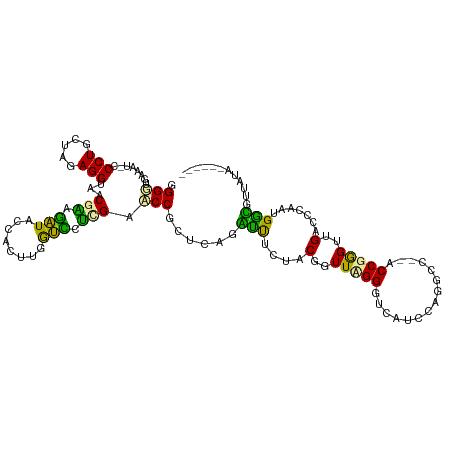
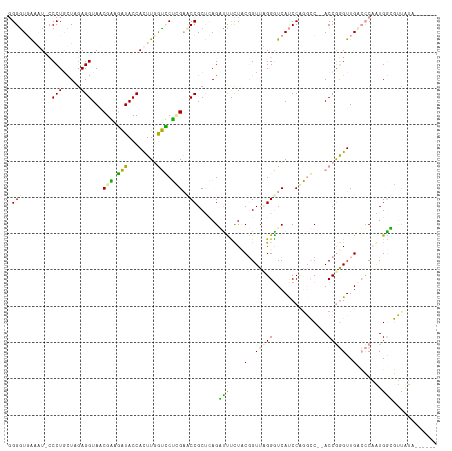
| Location | 169,975 – 170,093 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 60.45 |
| Shannon entropy | 0.57992 |
| G+C content | 0.56497 |
| Mean single sequence MFE | -47.70 |
| Consensus MFE | -16.37 |
| Energy contribution | -14.83 |
| Covariance contribution | -1.53 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
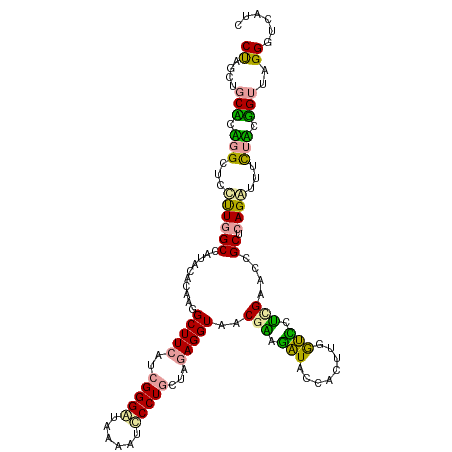
>dm3.chr3LHet 169975 118 - 2555491 CUAGCGGCUGUUGUGUCUUGGCCAGAGACAAAGCUUCUGCGGGUGACUUGGCCUGCUAAAGGUAACGAAGAUACCACUUCGUUCUGGCCCCGCUCAGCCCCUUACGGUUAGGGUCAUC .....(((((..(((....(((((((.......(((..((((((......))))))..)))...((((((......))))))))))))).))).)))))(((((....)))))..... ( -43.70, z-score = -1.40, R) >droVir3.scaffold_12823 182019 118 + 2474545 CUGGCUGCACAGGCUCCCUGGCCAUGCGCAAGGCUUCAUCGGGAUGAAAUCCCUUUUUGAGGUAGCGCACAUACCAUUUGGUGCGCGAUCCGCUCAGGUUUCUUCUGUUUGGAUCACC ...((.((((.((((....)))).(((((...((((((..(((((...)))))....)))))).)))))...........))))))((((((..((((.....))))..))))))... ( -46.50, z-score = -2.60, R) >droGri2.scaffold_4812 5895 118 - 6386 CCUCCUCCCUGGGCUCUUUGGCCUUUUUCAGGGCUUCAUCGGGGUUGAGUCCCUGCUGGAGGUACCGGAGGUACCACUUGACCCUUGAACCGCAGAGAUUUCUCCGGUUCGGGUCAUC (((((.(((((((((....)))).....)))))(((((.(((((......))))).))))).....))))).......((((((..((((((..(((....))))))))))))))).. ( -52.90, z-score = -2.67, R) >consensus CUAGCUGCACAGGCUCCUUGGCCAUACACAAGGCUUCAUCGGGAUAAAAUCCCUGCUAGAGGUAACGAAGAUACCACUUGGUCCUCGAACCGCUCAGAUUUCUACGGUUAGGGUCAUC ((....(((.(((...((((((..........(((((..(((((......)))))...)))))..(((.(((........))).)))....)).))))...))).)))..))...... (-16.37 = -14.83 + -1.53)

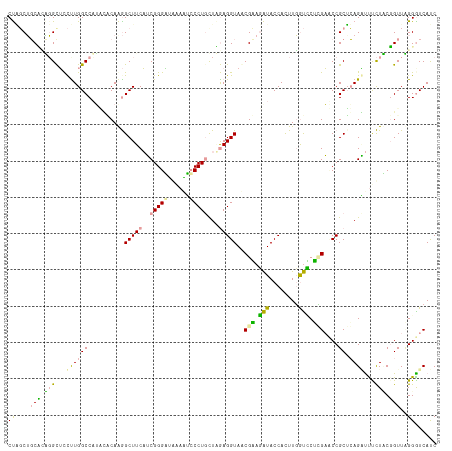
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:50:07 2011