| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,934,165 – 3,934,258 |
| Length | 93 |
| Max. P | 0.507916 |

| Location | 3,934,165 – 3,934,258 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.50 |
| Shannon entropy | 0.55246 |
| G+C content | 0.45241 |
| Mean single sequence MFE | -15.82 |
| Consensus MFE | -9.46 |
| Energy contribution | -9.21 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
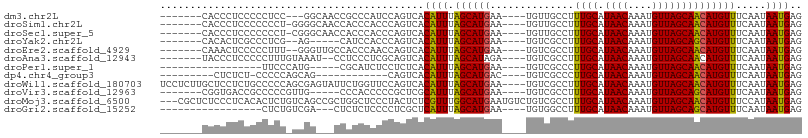
>dm3.chr2L 3934165 93 - 23011544 -------CACCCUCCCCCUCC---GGCAACCGCCCAUCCAGUCACAUUUAGCAUGAA----UGUUGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG -------..............---(((....)))..........((((........(----((((((....((((.....))))..)))))))........)))).. ( -16.19, z-score = -1.49, R) >droSim1.chr2L 3889149 95 - 22036055 -------CACCCUCCCCCCCU-GGGGCAACCACCCACCCAGUCACAUUUAGCAUGAA----UGUUGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG -------............((-(((...........)))))...((((........(----((((((....((((.....))))..)))))))........)))).. ( -19.99, z-score = -1.27, R) >droSec1.super_5 2037118 95 - 5866729 -------CACCCUCCCCCCCU-CGGGCAACCACCCACCCAGUCACAUUUAGCAUGAA----UGUUGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG -------..............-.((....)).............((((........(----((((((....((((.....))))..)))))))........)))).. ( -17.09, z-score = -1.40, R) >droYak2.chr2L 3948448 89 - 22324452 -------CACACUCGCCCUCG--AG-----CAUCCACCCAGUCACAUUUAGCAUGAA----UGUCGCCUUUGCAUAACAAAUGUUAGCAGCAUGUUUCAAUAAUGAG -------....((((.....(--((-----(((.(.....((.((((((.....)))----))).))...(((.((((....)))))))).))))))......)))) ( -14.90, z-score = -0.41, R) >droEre2.scaffold_4929 3992045 94 - 26641161 -------CAAACUCCCCCUUU--GGGUUGCCACCCAACCAGUCACAUUUAGCAUGAA----UGUCGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG -------....(((.....((--((((....))))))...((.((((((.....)))----))).))..((((.((((....))))))))..............))) ( -17.10, z-score = -0.55, R) >droAna3.scaffold_12943 480546 94 + 5039921 -------UACCCUCCCCCUUUGUAAAU--CCUCCCUCGCAGUCACAUUUAGCAUAGA----UGUCGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG -------....................--.....((((..((.(((((((...))))----))).))..((((.((((....)))))))).............)))) ( -11.60, z-score = -0.94, R) >droPer1.super_1 6716528 81 + 10282868 -----------------UUCCCAUG-----CGCAUCUCCUCUCACAUUUAGCAUGAA----UGUCGCCCUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG -----------------....((((-----(...................)))))..----((..((..((((.((((....))))))))...))..))........ ( -11.51, z-score = -0.24, R) >dp4.chr4_group3 9601659 81 + 11692001 ---------CUCUCU-CCCCCAGCAG------------CAGUCACAUUUAGCAUGAC----UGUCGCCCUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG ---------..(((.-......((.(------------((((((.........))))----))).))..((((.((((....))))))))..............))) ( -17.00, z-score = -2.11, R) >droWil1.scaffold_180703 1870783 103 - 3946847 UCCUCUUGCUCCUCUGCCCCCAGCGAGUAUUCUGGUUCCAGUCACAUUUAGCAUGAA----UGUCGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG ......(((((..(((....))).)))))..(((....)))...((((.((((((..----........((((.((((....)))))))))))))).....)))).. ( -17.60, z-score = -0.09, R) >droVir3.scaffold_12963 10857510 91 + 20206255 -------CGGUGACCGCCCCCGUUG-----CCCACCCCCGCUCGCAUUUAGCAUGAA----UGUCGCCUUUGCAUAACAAAUGUUAGCAGCAUGUUUCAAUAAUGAG -------.((..((.......))..-----))........(((((.....)).((((----....((...(((.((((....)))))))))....)))).....))) ( -16.40, z-score = 0.31, R) >droMoj3.scaffold_6500 1935834 104 - 32352404 ---CGCUCUCCCUCACACUCUGUCAGCCGCUGGCUCCCUACUCUCGUUUGGCAUGAAUGUCUGUCGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCCAUAAUGAG ---..................(((((...)))))........(((((((((((((...((.....))..((((.((((....))))))))))))...))).)))))) ( -16.90, z-score = 0.59, R) >droGri2.scaffold_15252 12458043 83 - 17193109 -----------------CUCUGUCGA---CUCUCUCCCCUCGCUCAUUUAGCAUGAA----UGUGGCCUUUGCAUAACAAAUGUUAGCAGCAUGUUUCAAUAAUGAG -----------------.........---.............((((((.((((((..----..((((.((((.....)))).))))....)))))).....)))))) ( -13.60, z-score = 0.23, R) >consensus _______CACCCUCCCCCUCU_UCGG___CCACCCACCCAGUCACAUUUAGCAUGAA____UGUCGCCUUUGCAUAACAAAUGUUAGCAACAUGUUUCAAUAAUGAG ............................................((((.((((((..............((((.((((....)))))))))))))).....)))).. ( -9.46 = -9.21 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:56 2011