
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 21,014,097 – 21,014,220 |
| Length | 123 |
| Max. P | 0.997551 |

| Location | 21,014,097 – 21,014,189 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.43425 |
| G+C content | 0.44506 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -11.48 |
| Energy contribution | -14.42 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
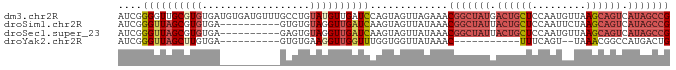
>dm3.chr2R 21014097 92 + 21146708 AUCGGGGUUGCGUGUGAUGUGAUGUUUGCCUGUAUGUUGAUCCAGUAGUUAGAAACGGCUAUGACUGCUCCAAUGUUAAGCAGUCAUAGCCG ....((((((((.(..(........)..).))).....)))))............((((((((((((((.........)))))))))))))) ( -30.70, z-score = -2.56, R) >droSim1.chr2R 19466552 82 + 19596830 AUCGGGUUAGCGUGUGA----------GUGUGUAGGUUGAUCAAGUAGUUAUAAACGGCUAUUACUGCUCCAAUUCUAAGCAGUCAUAGCCG ....(((((((.((...----------.....)).))))))).............(((((((.((((((.........)))))).))))))) ( -24.90, z-score = -2.52, R) >droSec1.super_23 870947 82 + 989336 AUCGGGUUAGCGUGUGA----------GAGUGUAGGUUGAUCAAGUAGUUAUAAACGGCUAUUACUGCUCCAAUGUUAAGCAGUCAUAGCCG ....(((((((.((...----------.....)).))))))).............(((((((.((((((.........)))))).))))))) ( -24.90, z-score = -2.53, R) >droYak2.chr2R 20984771 69 + 21139217 AUCGGGUUAGCUUGUGA----------GUGUGAAGGUUGGUUUGGUGGUUAUAAAC-----------UUUCAGU--UAAACGGCCAUGACUG ....((((((((.((..----------...((((((((...............)))-----------)))))..--...)))))..))))). ( -12.06, z-score = 0.64, R) >consensus AUCGGGUUAGCGUGUGA__________GUGUGUAGGUUGAUCAAGUAGUUAUAAACGGCUAUUACUGCUCCAAUGUUAAGCAGUCAUAGCCG ....((((((((((..................)))))))))).............(((((((.((((((.........)))))).))))))) (-11.48 = -14.42 + 2.94)
| Location | 21,014,097 – 21,014,189 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.43425 |
| G+C content | 0.44506 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -12.21 |
| Energy contribution | -14.15 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
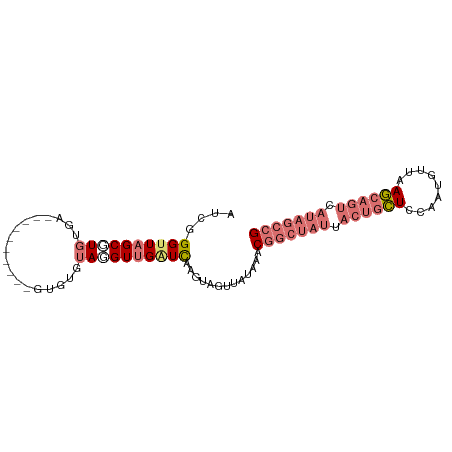
>dm3.chr2R 21014097 92 - 21146708 CGGCUAUGACUGCUUAACAUUGGAGCAGUCAUAGCCGUUUCUAACUACUGGAUCAACAUACAGGCAAACAUCACAUCACACGCAACCCCGAU (((((((((((((((.......)))))))))))))))..........(((..........)))............................. ( -28.50, z-score = -4.93, R) >droSim1.chr2R 19466552 82 - 19596830 CGGCUAUGACUGCUUAGAAUUGGAGCAGUAAUAGCCGUUUAUAACUACUUGAUCAACCUACACAC----------UCACACGCUAACCCGAU (((((((.(((((((.......))))))).)))))))............................----------................. ( -20.40, z-score = -3.37, R) >droSec1.super_23 870947 82 - 989336 CGGCUAUGACUGCUUAACAUUGGAGCAGUAAUAGCCGUUUAUAACUACUUGAUCAACCUACACUC----------UCACACGCUAACCCGAU (((((((.(((((((.......))))))).)))))))............................----------................. ( -20.40, z-score = -3.66, R) >droYak2.chr2R 20984771 69 - 21139217 CAGUCAUGGCCGUUUA--ACUGAAA-----------GUUUAUAACCACCAAACCAACCUUCACAC----------UCACAAGCUAACCCGAU ......(((..(((.(--((.....-----------)))...)))..)))...............----------................. ( -4.30, z-score = 0.71, R) >consensus CGGCUAUGACUGCUUAACAUUGGAGCAGUAAUAGCCGUUUAUAACUACUUGAUCAACCUACACAC__________UCACACGCUAACCCGAU (((((((.(((((((.......))))))).)))))))....................................................... (-12.21 = -14.15 + 1.94)
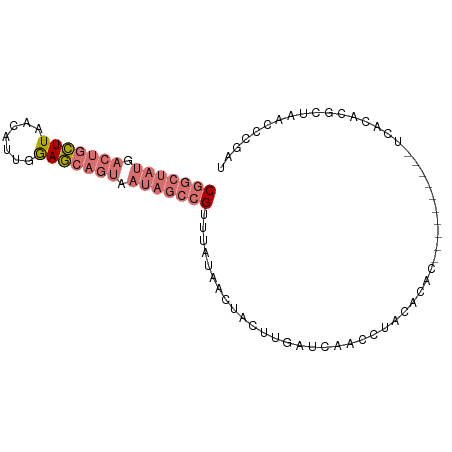

| Location | 21,014,128 – 21,014,220 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 51.32 |
| Shannon entropy | 0.92620 |
| G+C content | 0.40962 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -2.94 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
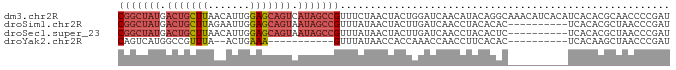
>dm3.chr2R 21014128 92 + 21146708 -----GUAUGUUGAUCCAGUAGUUAGAAACGGCUAUGACUGCUCCAAUGUUAAGCAGUCAUAGCCGUUUAUAGCCGGCUGUUGCAAUAAA-ACAGUUU- -----...(((((...((((.((((.(((((((((((((((((.........))))))))))))))))).))))..))))...)))))..-.......- ( -39.90, z-score = -7.14, R) >droSim1.chr2R 19466573 92 + 19596830 -----GUAGGUUGAUCAAGUAGUUAUAAACGGCUAUUACUGCUCCAAUUCUAAGCAGUCAUAGCCGUUUAUAGCCGGCUGUUACAAUAAU-ACAGUUU- -----(((.((((....(((.(((((((((((((((.((((((.........)))))).)))))))))))))))..)))....))))..)-)).....- ( -35.10, z-score = -6.35, R) >droSec1.super_23 870968 92 + 989336 -----GUAGGUUGAUCAAGUAGUUAUAAACGGCUAUUACUGCUCCAAUGUUAAGCAGUCAUAGCCGUUUAUAGCCGGCUGUUACAAUAAU-ACAGUUU- -----(((.((((....(((.(((((((((((((((.((((((.........)))))).)))))))))))))))..)))....))))..)-)).....- ( -35.10, z-score = -5.95, R) >droYak2.chr2R 20984792 86 + 21139217 -----GAAGGUUGGUUUGGUGGUUAUAAAC-----UUUCAGUUAAACGGCCAUGACUGCUUAGCUAUUGGU-GUUGGCUAUUACAAUAAA-UCAGCUU- -----..((((((((((((((((((...((-----(...((((((.(((......))).))))))...)))-..))))))).....))))-)))))))- ( -22.10, z-score = -1.44, R) >droEre2.scaffold_4845 22432301 91 + 22589142 GGUGUGUUGGUCGAUCAGGUAGUUAUAAAC-----UUUCAGUUAAGCGGCCAUGACUGCUUAGCGAUUGGU-GUUGGCCAUUACAAUAAA-UCAGCUU- ..((((.((((((((..((.(((.....))-----).)).(((((((((......))))))))).......-)))))))).)))).....-.......- ( -25.40, z-score = -1.39, R) >droWil1.scaffold_180700 5286491 80 - 6630534 ---------GUGGGAUGACUGACUCAAGGU-----UGUGGCUUAUAAAUUUGCCAAGAGAUUGC-----CUUUCGGUCAAUUAUAUUAAAGCCAGCUGA ---------......(((((((...(((((-----..((((..........)))).......))-----)))))))))).................... ( -17.60, z-score = -0.02, R) >droMoj3.scaffold_6496 22981177 69 + 26866924 ------------------GUGAG-CAGCAU-----GUGGGGUUAUAAAUUCAGUGCGAAAUUGCA----CUGGUCGCCCAUUACAGUAAC-UCAGCUG- ------------------.((((-..((.(-----(((((((.......((((((((....))))----))))..))))..))))))..)-)))....- ( -21.00, z-score = -1.47, R) >droGri2.scaffold_15112 781693 70 - 5172618 --------------AUGCCUGGUUAUAAAU--------UAGCUGGAAAAUUGCCUUGGCUUAGAC-----CAGUUGGCCAUUAUACUAAA-UCAGCUG- --------------..((.(((((((((.(--------(((((((......((....)).....)-----)))))))...)))))....)-)))))..- ( -14.20, z-score = 0.27, R) >consensus _____GUAGGUUGAUCAAGUAGUUAUAAAC_____UUACAGCUACAAAGUUAAGAAGGCAUAGCC_UU_AUAGCCGGCUAUUACAAUAAA_UCAGCUU_ ................................................................................................... ( 0.00 = 0.00 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:54 2011