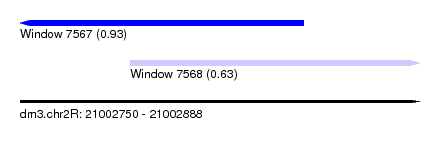
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 21,002,750 – 21,002,888 |
| Length | 138 |
| Max. P | 0.926874 |

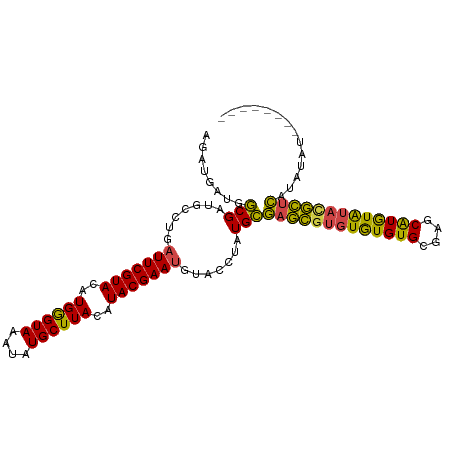
| Location | 21,002,750 – 21,002,848 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Shannon entropy | 0.28999 |
| G+C content | 0.42960 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -26.87 |
| Energy contribution | -25.66 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 21002750 98 - 21146708 AGAUGAUGGCGAUGCCUGAUUCGUACAUGGGUAAAUAUGCUUACAUACGAAUGUACCUUUGCGAGCGUGUGUGUGUGAGCAUGUAUACGCUCAUAUAU-------- ........((((......(((((((..((((((....))))))..)))))))......))))(((((((((..((....))..)))))))))......-------- ( -32.90, z-score = -2.22, R) >droSim1.chr2R 19454972 98 - 19596830 AGAUGAUGGCGAUGCCUGAUUCGUACAUGGGUAAAUAUGCUUACAUACGAAUGUACCUUUGCGAGCGUGUGUGUGCGAGCAUGUAUAUGCUCAUAUAU-------- ........((((......(((((((..((((((....))))))..)))))))......))))(((((((((..((....))..)))))))))......-------- ( -31.30, z-score = -1.59, R) >droSec1.super_23 859406 98 - 989336 AGAUGAUGGCGAUGCCUGAUUCGUACAUGGGUAAAUAUGCUUACAUACGAAUGUACCUUUGCGAGCGUGUGUGUGCGAGCAUGUAUAUGCUCAUAUAU-------- ........((((......(((((((..((((((....))))))..)))))))......))))(((((((((..((....))..)))))))))......-------- ( -31.30, z-score = -1.59, R) >droYak2.chr2R 20973143 98 - 21139217 AGAUGAUGGCGAUGCCUGAUUCGUACAUGGGUAAAUAUGCUUACAUACGAAUGUACCUAUGCGAGCGUGUGUGUGCGAGCAUAUAUACGCUCAUAUAU-------- ..((((.(((...)))....)))).((((((((..((((....))))......)))))))).(((((((((((((....)))))))))))))......-------- ( -34.20, z-score = -2.49, R) >droEre2.scaffold_4845 22412208 98 - 22589142 AGUUGAUGGCGAUGCCUGAUUCGUACAUGGGUAAAUAUGCUUACAUACGAAUGUACCUAUGCGAGCGUGUGUGUGCGAGCAUGUAUACGCUCAUAUAU-------- .......(((...)))..(((((((..((((((....))))))..))))))).....((((.(((((((((..((....))..))))))))).)))).-------- ( -32.00, z-score = -1.35, R) >droAna3.scaffold_13266 12309858 106 - 19884421 GGAGGAUGACGGUGCCUGAUUCGUACAUGAGUAAAUAUGCUUACAUACGAACGAAUGCAUGUACAUACCUAUGUGAAGACGUAUGUACAUAUGUACGGACGUGUCU ..(((((((((.(((.((.((((((..((((((....))))))..))))))))...)))))).))).)))....((..((((.(((((....))))).)))).)). ( -28.70, z-score = -1.02, R) >consensus AGAUGAUGGCGAUGCCUGAUUCGUACAUGGGUAAAUAUGCUUACAUACGAAUGUACCUAUGCGAGCGUGUGUGUGCGAGCAUGUAUACGCUCAUAUAU________ ........(((.......(((((((..((((((....))))))..))))))).......)))(((((((((((((....))))))))))))).............. (-26.87 = -25.66 + -1.21)

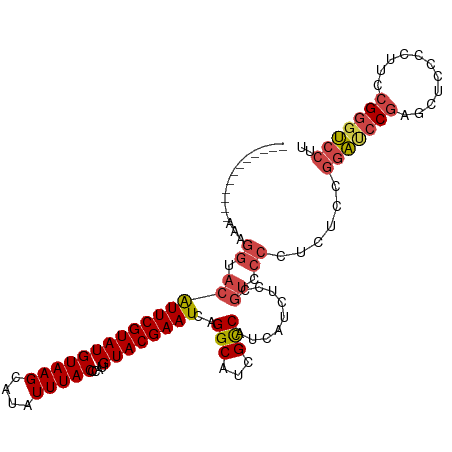
| Location | 21,002,788 – 21,002,888 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.68 |
| Shannon entropy | 0.27689 |
| G+C content | 0.52309 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 21002788 100 + 21146708 ------------AAAGGUACAUUCGUAUGUAAGCAUAUUUACCCAUGUACGAAUCAGGCAUCGCCAUCAUCUCCGUCCCCUCACCGGAUCCGAGCUCCCCUUCCGGGUCCUU ------------...(((..(((((((((((((....)))))....))))))))..(((...))).................)))(((((((...........))))))).. ( -23.90, z-score = -1.73, R) >droSim1.chr2R 19455010 100 + 19596830 ------------AAAGGUACAUUCGUAUGUAAGCAUAUUUACCCAUGUACGAAUCAGGCAUCGCCAUCAUCUCCGUCCCCUCUCCGGAUCCGAGCUCCCCUUCCGGGUCCGU ------------...((.(((((((((((((((....)))))....))))))))..(((...))).........)).)).....((((((((...........)))))))). ( -25.20, z-score = -1.91, R) >droSec1.super_23 859444 100 + 989336 ------------AAAGGUACAUUCGUAUGUAAGCAUAUUUACCCAUGUACGAAUCAGGCAUCGCCAUCAUCUCCGUCCCCUCUCCGGAUCCGAGCUCCCUUUCCGGGUCCGU ------------...((.(((((((((((((((....)))))....))))))))..(((...))).........)).)).....((((((((...........)))))))). ( -25.20, z-score = -1.97, R) >droYak2.chr2R 20973181 100 + 21139217 ------------AUAGGUACAUUCGUAUGUAAGCAUAUUUACCCAUGUACGAAUCAGGCAUCGCCAUCAUCUCCGUCCCCUCCCCGGAUCCGAGCUCCCCUUCCGGCUCCUU ------------...((.(((((((((((((((....)))))....))))))))..(((...))).........)).))......(((.(((...........))).))).. ( -19.40, z-score = -0.69, R) >droEre2.scaffold_4845 22412246 100 + 22589142 ------------AUAGGUACAUUCGUAUGUAAGCAUAUUUACCCAUGUACGAAUCAGGCAUCGCCAUCAACUCCGUCCCCUCUCCGGAUCCGAGCUCCCCAUCCGGGUCCUU ------------...((.(((((((((((((((....)))))....))))))))..(((...))).........)).))......(((((((...........))))))).. ( -23.40, z-score = -1.39, R) >droAna3.scaffold_13266 12309892 108 + 19884421 AUAGGUAUGUACAUGCAUUCGUUCGUAUGUAAGCAUAUUUACUCAUGUACGAAUCAGGCACCGUCAUCCUCCCCACCCCCUAU----GCCCCACCGCCCCUACCGAGCUCCU ...((((((((((((.....((..(((((....)))))..)).)))))))).....((((......................)----)))..........))))........ ( -18.65, z-score = -0.69, R) >consensus ____________AAAGGUACAUUCGUAUGUAAGCAUAUUUACCCAUGUACGAAUCAGGCAUCGCCAUCAUCUCCGUCCCCUCUCCGGAUCCGAGCUCCCCUUCCGGGUCCUU ...............((.(((((((((((((((....)))))....))))))))..(((...))).........)).))......(((((((...........))))))).. (-17.76 = -18.60 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:52 2011