| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,997,453 – 20,997,553 |
| Length | 100 |
| Max. P | 0.938925 |

| Location | 20,997,453 – 20,997,553 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.63 |
| Shannon entropy | 0.46288 |
| G+C content | 0.39206 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -15.26 |
| Energy contribution | -14.56 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
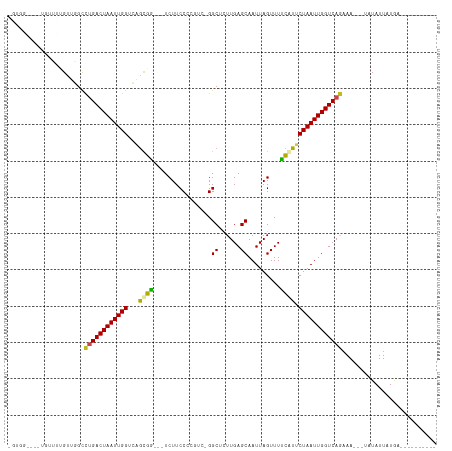
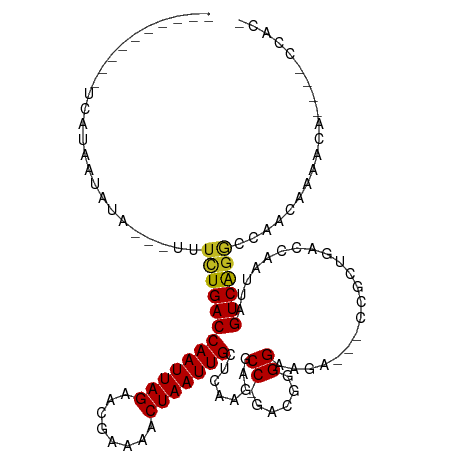
>dm3.chr2R 20997453 100 + 21146708 -GUGGAUUAUGUUUUGUUGGUCUGACUAAUUGGUCUACAG---UCUUCCCCGUC-GGCUCUUGAGCAAUUAGUUUUUAUUCUAAUUGGUCAGAAA---UAUAUUAUGA---------- -......((((((((...(..(((((.....((.(....)---.)).....)))-))..).(((.(((((((........))))))).)))))))---))))......---------- ( -20.60, z-score = -0.73, R) >droSim1.chr2R 19449570 100 + 19596830 -GUGGAUUAUGUUUUGUUGGUCUGACUAAUUGGUCAACAG---UCUUCCCCGUC-GGCUCUUGAGCAAUUAGUUUUUAUUCUAAUUGGUCAGAAA---UAUAUUAUGA---------- -......((((((((..((..(.(((((((((.((((.((---((.........-)))).)))).)))))))))............)..))))))---))))......---------- ( -23.22, z-score = -1.58, R) >droSec1.super_23 854007 100 + 989336 -GUGGAUUAUGUUUUGUUGGUCUGACUAAUUGGUCAGCAG---UCUUCCCCGUC-GGCUCUUGAGCAAUUAGUUUUUAUUCUAAUUGGUCAGAAA---UAUAUUAUGA---------- -......((((((((..((..(.(((((((((.((((.((---((.........-)))).)))).)))))))))............)..))))))---))))......---------- ( -22.92, z-score = -1.25, R) >droYak2.chr2R 20967544 100 + 21139217 -GGGGAUAAUGUUUUGUUGGCCUGACUAAUUGGUCAGUAG---UCUUCCCCGUC-GGCUCUUGAGCAAUUAGUUUUUAUUCUAAUUGGUCAGAAA---UAUAUUAUGA---------- -....(((((((....((((((.(((((((((.((((.((---((.........-)))).)))).)))))))))............))))))...---.)))))))..---------- ( -27.82, z-score = -2.29, R) >droEre2.scaffold_4845 22406438 100 + 22589142 -GUGGAUAAUGUUUUGUUGGCCUGACUAAUUGGUCAGUAG---UCUUCCCCGUC-GGCUCUUGAGCAAUUAGUUUUUAUUCUAAUUGGUCAGAAA---UAUAUUAUGA---------- -....(((((((....((((((.(((((((((.((((.((---((.........-)))).)))).)))))))))............))))))...---.)))))))..---------- ( -27.82, z-score = -2.94, R) >droAna3.scaffold_13266 12304537 100 + 19884421 -GUGGCCUCUGUUUUGCUGGCCUGACUAAUUGGUCAGCGG---UCUUCCC-CGUCGGCUCUUGAGCAAUUAGUUUUCGUUCUAAUUGGUCAGAAA---UAUAUUAUGA---------- -..((((........(((((((.........)))))))((---......)-)...)))).((((.(((((((........))))))).))))...---..........---------- ( -29.40, z-score = -2.26, R) >dp4.chr3 10107123 96 + 19779522 ----------AUGUGGCUCGCCCGACUAAUUGGUCAGCGGCGGUCUUCCAACGUCGGCUCUUGAGCAAUUAGUUUUCGUUCUAAUUGGUCAGAAAA--UAUAUUAUGA---------- ----------....(((.(((((((((....)))).).))))))).............(((.((.(((((((........))))))).)))))...--..........---------- ( -24.40, z-score = -0.66, R) >droPer1.super_4 5436449 96 + 7162766 ----------AUGUGGCGCGCCCGACUAAUUGGUCAGCGGCGGUCUUCCAACGUCGGCUCUUGAGCAAUUAGUUUUCGUUCUAAUUGGUCAGAAAA--UAUAUUAUGA---------- ----------...(((((.((((((((....)))).).)))((....))..)))))..(((.((.(((((((........))))))).)))))...--..........---------- ( -25.10, z-score = -0.41, R) >droWil1.scaffold_180700 5258739 113 - 6630534 UCUAUUAAUGUUUGUGUUGGUCUGACUAAUUGGUCUGCGA---UCUUCCCGUU--GGCUUUUGAGCAAUUAGUUUUCAUUCUAAUUGGUCAACAAAAAUUUAUUAUAAAGAGAAACUG (((..(((((..(.(((((..(.(((((((((.((.((.(---.(.....).)--.))....)).)))))))))............)..)))))...)..)))))...)))....... ( -22.52, z-score = -1.22, R) >droVir3.scaffold_12875 18312336 85 - 20611582 --------------UGUCGGCCUGACUAAUUGGAUGGCGU---UCUUCCAGUU--GGCGCUUGAGCAAUUAGUUUACGUUCUAAUUGGUCAGAAA----UUAUUAUAA---------- --------------.......(((((((((((((..((((---....(.((((--(.(......)))))).)...)))))))))))))))))...----.........---------- ( -23.30, z-score = -1.91, R) >droMoj3.scaffold_6496 22952274 82 + 26866924 -----------------UGCUCUGACUAAUUGGACGGCGU---UCUUCCAGUU--GGCGCUUGAGCAAUUAGUUUACGUUCUAAUUGGUCAGAAA----UUAUUAUAA---------- -----------------...((((((((((((((((((((---(.........--))))).(((((.....))))))).))))))))))))))..----.........---------- ( -26.20, z-score = -3.64, R) >droGri2.scaffold_15112 751678 87 - 5172618 --------------UGUCGGCUUGACUAAUUGGACGGCGU---UCUUCCAGUUUUGGCGCUUGAGCAAUUAGUUUACGUUCUAAUUGGUCAGAAA----UUAUUAUAA---------- --------------.......(((((((((((((((((((---(...........))))).(((((.....))))))).)))))))))))))...----.........---------- ( -22.10, z-score = -1.59, R) >consensus _GUGG____UGUUUUGUUGGCCUGACUAAUUGGUCAGCGG___UCUUCCCCGUC_GGCUCUUGAGCAAUUAGUUUUCAUUCUAAUUGGUCAGAAA___UAUAUUAUGA__________ .....................((((((((((((...((((....((..........((......))....))...)))).)))))))))))).......................... (-15.26 = -14.56 + -0.71)


| Location | 20,997,453 – 20,997,553 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.63 |
| Shannon entropy | 0.46288 |
| G+C content | 0.39206 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -11.00 |
| Energy contribution | -10.77 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
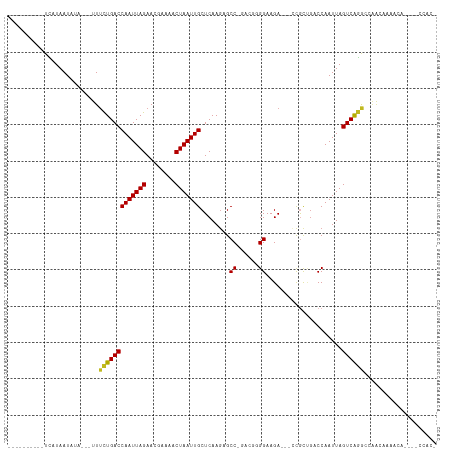
>dm3.chr2R 20997453 100 - 21146708 ----------UCAUAAUAUA---UUUCUGACCAAUUAGAAUAAAAACUAAUUGCUCAAGAGCC-GACGGGGAAGA---CUGUAGACCAAUUAGUCAGACCAACAAAACAUAAUCCAC- ----------..........---..(((((((((((((........)))))))..........-.((((......---))))..........))))))...................- ( -15.70, z-score = -1.48, R) >droSim1.chr2R 19449570 100 - 19596830 ----------UCAUAAUAUA---UUUCUGACCAAUUAGAAUAAAAACUAAUUGCUCAAGAGCC-GACGGGGAAGA---CUGUUGACCAAUUAGUCAGACCAACAAAACAUAAUCCAC- ----------..........---..(((((((((((((........))))))).........(-(((((......---))))))........))))))...................- ( -19.20, z-score = -2.55, R) >droSec1.super_23 854007 100 - 989336 ----------UCAUAAUAUA---UUUCUGACCAAUUAGAAUAAAAACUAAUUGCUCAAGAGCC-GACGGGGAAGA---CUGCUGACCAAUUAGUCAGACCAACAAAACAUAAUCCAC- ----------..........---..(((((((((((((........)))))))..........-....((..((.---...))..)).....))))))...................- ( -16.10, z-score = -1.57, R) >droYak2.chr2R 20967544 100 - 21139217 ----------UCAUAAUAUA---UUUCUGACCAAUUAGAAUAAAAACUAAUUGCUCAAGAGCC-GACGGGGAAGA---CUACUGACCAAUUAGUCAGGCCAACAAAACAUUAUCCCC- ----------..........---.((((((.(((((((........))))))).)).))))..-...(((((...---...(((((......)))))(....).........)))))- ( -20.80, z-score = -2.76, R) >droEre2.scaffold_4845 22406438 100 - 22589142 ----------UCAUAAUAUA---UUUCUGACCAAUUAGAAUAAAAACUAAUUGCUCAAGAGCC-GACGGGGAAGA---CUACUGACCAAUUAGUCAGGCCAACAAAACAUUAUCCAC- ----------..(((((...---..(((((.(((((((........))))))).)).)))(((-(((.((..((.---...))..)).....))).))).........)))))....- ( -19.60, z-score = -2.64, R) >droAna3.scaffold_13266 12304537 100 - 19884421 ----------UCAUAAUAUA---UUUCUGACCAAUUAGAACGAAAACUAAUUGCUCAAGAGCCGACG-GGGAAGA---CCGCUGACCAAUUAGUCAGGCCAGCAAAACAGAGGCCAC- ----------..........---..(((((.(((((((........))))))).)).)))(((...(-(......---))((((.((.........)).))))........)))...- ( -23.40, z-score = -2.21, R) >dp4.chr3 10107123 96 - 19779522 ----------UCAUAAUAUA--UUUUCUGACCAAUUAGAACGAAAACUAAUUGCUCAAGAGCCGACGUUGGAAGACCGCCGCUGACCAAUUAGUCGGGCGAGCCACAU---------- ----------..........--(((((..((.((((((........))))))(((....)))....))..)))))..(((((((((......))).)))).)).....---------- ( -21.60, z-score = -1.47, R) >droPer1.super_4 5436449 96 - 7162766 ----------UCAUAAUAUA--UUUUCUGACCAAUUAGAACGAAAACUAAUUGCUCAAGAGCCGACGUUGGAAGACCGCCGCUGACCAAUUAGUCGGGCGCGCCACAU---------- ----------..........--(((((..((.((((((........))))))(((....)))....))..)))))..(((((((((......))).)))).)).....---------- ( -21.60, z-score = -1.11, R) >droWil1.scaffold_180700 5258739 113 + 6630534 CAGUUUCUCUUUAUAAUAAAUUUUUGUUGACCAAUUAGAAUGAAAACUAAUUGCUCAAAAGCC--AACGGGAAGA---UCGCAGACCAAUUAGUCAGACCAACACAAACAUUAAUAGA ..(((..(((..........(((((((((...((((((........))))))(((....))))--))))))))..---.....(((......))))))..)))............... ( -14.80, z-score = -0.06, R) >droVir3.scaffold_12875 18312336 85 + 20611582 ----------UUAUAAUAA----UUUCUGACCAAUUAGAACGUAAACUAAUUGCUCAAGCGCC--AACUGGAAGA---ACGCCAUCCAAUUAGUCAGGCCGACA-------------- ----------.........----..(((((((((((((........)))))))..........--...((((.(.---....).))))....))))))......-------------- ( -14.50, z-score = -1.01, R) >droMoj3.scaffold_6496 22952274 82 - 26866924 ----------UUAUAAUAA----UUUCUGACCAAUUAGAACGUAAACUAAUUGCUCAAGCGCC--AACUGGAAGA---ACGCCGUCCAAUUAGUCAGAGCA----------------- ----------.........----.((((((((((((((........)))))))..........--...((((.(.---....).))))....)))))))..----------------- ( -16.00, z-score = -1.77, R) >droGri2.scaffold_15112 751678 87 + 5172618 ----------UUAUAAUAA----UUUCUGACCAAUUAGAACGUAAACUAAUUGCUCAAGCGCCAAAACUGGAAGA---ACGCCGUCCAAUUAGUCAAGCCGACA-------------- ----------......(((----((..(((.(((((((........))))))).))).((((((....)))....---.))).....)))))(((.....))).-------------- ( -13.30, z-score = -0.76, R) >consensus __________UCAUAAUAUA___UUUCUGACCAAUUAGAACGAAAACUAAUUGCUCAAGAGCC_GACGGGGAAGA___CCGCUGACCAAUUAGUCAGGCCAACAAAACA____CCAC_ .........................(((((((((((((........)))))))........((......)).....................)))))).................... (-11.00 = -10.77 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:50 2011