| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,982,017 – 20,982,121 |
| Length | 104 |
| Max. P | 0.770507 |

| Location | 20,982,017 – 20,982,121 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.28 |
| Shannon entropy | 0.52742 |
| G+C content | 0.65041 |
| Mean single sequence MFE | -44.19 |
| Consensus MFE | -17.37 |
| Energy contribution | -18.55 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
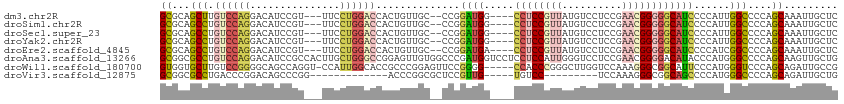
>dm3.chr2R 20982017 104 + 21146708 GCGCAGCUUGUCCAGGACAUCCGU---UUCCUGGACCACUGUUGC--CCGGAUGG----CCUCCGUUAUGUCCUCCGAACGGGGGCAUCCCCAUUGGCCCCAGCAAAUUGCUC (((((((..((((((((.......---.))))))))....)))))--..(((((.----((((((((.((.....)))))))))))))))......))...(((.....))). ( -42.30, z-score = -2.60, R) >droSim1.chr2R 19433824 104 + 19596830 GCGCAGCCUGUCCAGGACAUCCGU---UUCCUGGACCACUGUUGC--CCGGAUGG----CCUCCGUUAUGUCCUCCGAACGGGGGCAUCCCCAUUGGCCCCAGCAAAUUGCUC (((((((..((((((((.......---.))))))))....)))))--..(((((.----((((((((.((.....)))))))))))))))......))...(((.....))). ( -41.60, z-score = -2.29, R) >droSec1.super_23 838260 104 + 989336 GCGCAGCCUGUCCAGGACAUCCGU---UUCCUGGACCACUGUUGC--CCGGAUGG----CCUCCGUUAUGUCCUCCGAACGGGGGCAUCCCCAUUGGCCCCAGCAAAUUGCUC (((((((..((((((((.......---.))))))))....)))))--..(((((.----((((((((.((.....)))))))))))))))......))...(((.....))). ( -41.60, z-score = -2.29, R) >droYak2.chr2R 20952046 104 + 21139217 GCGCAGCCUGUCCAGGACAUCCGU---UUCCUGGACCACUGUUGC--CCGGAUGG----CCUCCGUUAUGUCCUCCGAACGGGGGCAUCCCCAUUGGCCCCAGCAAAUUGCUC (((((((..((((((((.......---.))))))))....)))))--..(((((.----((((((((.((.....)))))))))))))))......))...(((.....))). ( -41.60, z-score = -2.29, R) >droEre2.scaffold_4845 22390684 104 + 22589142 GCGCAGCCUGUCCAGGACAUCCGU---UUCCUGGACCACUGUUGC--CCGGAUGA----CCUCCGUUAUGUCCUCCGAACGGGGGCAUCCCCAUCGGCCCCAGCAAAUUGCUC (((((((..((((((((.......---.))))))))....)))))--..(((((.----((((((((.((.....)))))))))))))))......))...(((.....))). ( -41.60, z-score = -2.85, R) >droAna3.scaffold_13266 12289268 113 + 19884421 GCGGCGCCUGUCCAGGACAUCCGCCACUUGCUGGGCCGGAGUUGUGGCCCGAUGGUCCUCCUCCAUUGGGUCCUCCGAACGGGGACAUACCCAUGGGCCCCAGCAAGUUGCUG ((((((..((((...))))..))))((((((((((((((((....((((((((((.......))))))))))))))....(((......)))...)).)))))))))).)).. ( -60.80, z-score = -3.82, R) >droWil1.scaffold_180700 5236043 107 - 6630534 GUGGUGCUUGUCCGGGGCAGCCAGGU-CCAUUGGCACCGCCCGGAGUUCCGGGG-----CCACCCGGGCUUGGUCCAAAGGGCGGCAUUCCCAUGGGUCCCAGCAGAUUGCCG .........((((..((..(((((((-((..((((....(((((....))))))-----)))...)))))))))))...))))((((.((...(((...)))...)).)))). ( -47.90, z-score = 0.42, R) >droVir3.scaffold_12875 18288179 86 - 20611582 GCGGCGCCUGACCCGGACAGCCCGG-------------ACCCGGCGCUCCGUUG-----UGUCC---------UCCAAAGGGCGGCAGCCCCAUGGGCCCCAGCAGAUUGCUG ((((((((.(..((((.....))))-------------..).))))))..((((-----.(..(---------.(((..((((....))))..))))..))))).....)).. ( -36.10, z-score = -0.01, R) >consensus GCGCAGCCUGUCCAGGACAUCCGU___UUCCUGGACCACUGUUGC__CCGGAUGG____CCUCCGUUAUGUCCUCCGAACGGGGGCAUCCCCAUUGGCCCCAGCAAAUUGCUC ((...(((.((((((...............))))))..............((((.....((((((((..........))))))))))))......)))....))......... (-17.37 = -18.55 + 1.17)
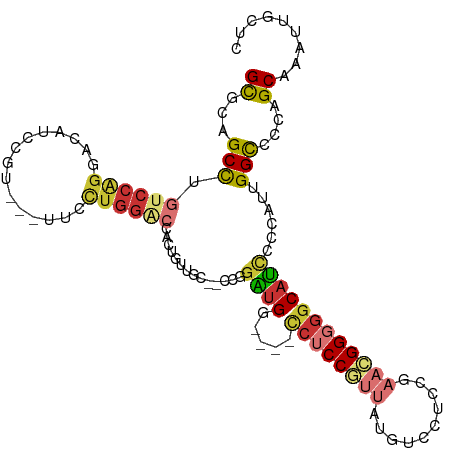

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:47 2011