| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,974,500 – 20,974,601 |
| Length | 101 |
| Max. P | 0.987668 |

| Location | 20,974,500 – 20,974,601 |
|---|---|
| Length | 101 |
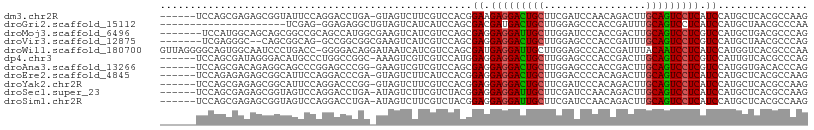
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.48 |
| Shannon entropy | 0.56016 |
| G+C content | 0.59321 |
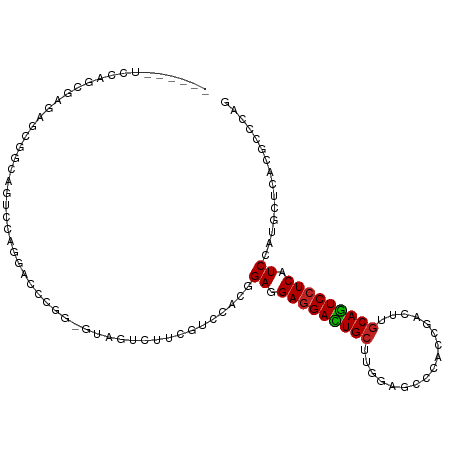
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.11 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20974500 101 + 21146708 ------UCCAGCGAGAGCGGUAUUCCAGGACCUGA-GUAGUCUUCGUCCACGGAAGAGGACUGCUUCGAUCCAACAGACUUGCAGUCCUCAUCCAUGCUCACGCCAAG ------....(((.((((((....)).((((..((-(....))).))))..(((.(((((((((.((.........))...))))))))).)))..)))).))).... ( -40.40, z-score = -4.01, R) >droGri2.scaffold_15112 717726 86 - 5172618 ---------------------UCGAG-GGAGAGGCUGUAGUCAUCAUCCAGCGACGAUGACUGCUUGGAGCCCACCGAUUUGCAGUCCUCAUCCAUGCUAACGCCCAA ---------------------.....-((((((((((((((((((.((....)).))))))...((((......))))..))))).)))).)))..((....)).... ( -28.80, z-score = -1.62, R) >droMoj3.scaffold_6496 22916091 101 + 26866924 -------UCCAUGGCAGCAGCGGCCGCAGCCAUGGCGAAGUCAUCGUCCAGCGAGGAGGAUUGCUUGGAUCCCACCGACUUGCAGUCCUCGUCCAUGCUGACGCCCAG -------.....(((..(((((((....))).((((((.....))).)))(.((.(((((((((((((......))))...))))))))).)))..))))..)))... ( -41.00, z-score = -1.76, R) >droVir3.scaffold_12875 18278727 98 - 20611582 -------UCGAGGGC--CAGCGGCAG-GCCGGCGGCGAAGUCAUCGUCCAGCGAGGAGGACUGCUUGGAGCCCACCGAUUUGCAGUCCUCGUCCAUGCUAACGCCCAG -------....((((--.((((((.(-(.((..(((...)))..)).)).))((.(((((((((((((......))))...))))))))).))..))))...)))).. ( -42.10, z-score = -1.33, R) >droWil1.scaffold_180700 5229192 107 - 6630534 GUUAGGGGCAGUGGCAAUCCCUGACC-GGGGACAGGAUAAUCAUCGUCCAGCGAUGAGGAUUGCUUGGAGCCCACCGAUUUACAAUCCUCAUCCAUGGUCACGCCCAA .....((((.(((((..(((((....-)))))((((((.......)))).(.(((((((((((.((((......))))....)))))))))))).))))))))))).. ( -45.00, z-score = -3.13, R) >dp4.chr3 10086400 101 + 19779522 ------UCCAGCGAUAGGGACAUGCCCUGGCCGGC-AAAGUCGUCGUCCAUGGAGGAGGACUGCUUGGAGCCCACCGACUUGCAGUCCUCGUCCAUUGUCACGCCCAG ------....(((..((((.....))))((.((((-......)))).))(((((.(((((((((((((......))))...))))))))).))))).....))).... ( -43.10, z-score = -2.44, R) >droAna3.scaffold_13266 12283654 101 + 19884421 ------UCCAGCGACAGAGGCAGCCCGGAGCCCGG-GAAGUCGUCGUCCAGCGAGGAGGACUGCUUGGAGCCCACCGACUUGCAGUCCUCGUCCAUGGUGACACCCAG ------............(((..(((((...))))-)..)))((((.(((..((.(((((((((((((......))))...))))))))).))..)))))))...... ( -43.60, z-score = -1.93, R) >droEre2.scaffold_4845 22384509 101 + 22589142 ------UCCAGAGAGAGCGGCAUUCCAGGACCCGA-GUAGUCUUCAUCCACGGAGGAGGACUGCUUGGACCCCACAGACUUGCAGUCCUCAUCCAUGCUCACGCCAAG ------........((((((....)).(((.((((-(((((((((.((....)).)))))))))))))........(((.....)))....)))..))))........ ( -35.50, z-score = -2.00, R) >droYak2.chr2R 20945899 101 + 21139217 ------UCCAGCGAGAGCGGCAUUCCAGGACCCGG-GUAGUCUUCGUCCACGGAGGAGGACUGCUUCGAUCCCACAGACUUGCAGUCCUCAUCCAUGCUCACGCCAAG ------....(((.(((((........((((..((-.....))..))))..(((.(((((((((.((.........))...))))))))).))).))))).))).... ( -39.10, z-score = -2.51, R) >droSec1.super_23 832489 101 + 989336 ------UCCAGCGAGAGCGGUAGUCCAGGACCUGA-AUAGUCUUCGUCUACGGAGGAGGAUUGCUUCGAUCCAACAGACUUGCAGUCCUCAUCCAUGCUCACGCCAAG ------....(((.(((((((((.(.(((((....-...))))).).))))(((.(((((((((.((.........))...))))))))).))).))))).))).... ( -40.80, z-score = -3.80, R) >droSim1.chr2R 19427980 101 + 19596830 ------UCCAGCGAGAGCGGUAGUCCAGGACCUGA-AUAGUCUUCGUCUACGGAGGAGGAUUGCUUCGAUCCAACAGACUUGCAGUCCUCAUCCAUGCUCACGCCAAG ------....(((.(((((((((.(.(((((....-...))))).).))))(((.(((((((((.((.........))...))))))))).))).))))).))).... ( -40.80, z-score = -3.80, R) >consensus ______UCCAGCGAGAGCGGCAGUCCAGGACCCGG_GUAGUCUUCGUCCACGGAGGAGGACUGCUUGGAGCCCACCGACUUGCAGUCCUCAUCCAUGCUCACGCCCAG ....................................................((.(((((((((.................))))))))).))............... (-16.24 = -16.11 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:46 2011