
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,964,287 – 20,964,362 |
| Length | 75 |
| Max. P | 0.984187 |

| Location | 20,964,287 – 20,964,362 |
|---|---|
| Length | 75 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 89.53 |
| Shannon entropy | 0.23596 |
| G+C content | 0.60900 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -31.23 |
| Energy contribution | -30.93 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
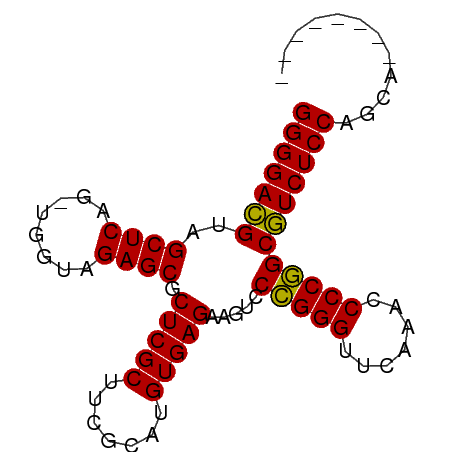
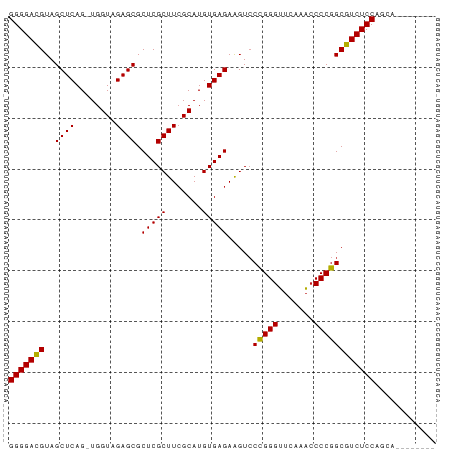
>dm3.chr2R 20964287 75 + 21146708 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAACA-------- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))....-------- ( -30.90, z-score = -2.05, R) >droGri2.scaffold_15112 678172 81 - 5172618 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAAUUGCUACG-- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))..........-- ( -30.90, z-score = -1.56, R) >droMoj3.scaffold_6496 22869371 75 + 26866924 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAGCG-------- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))....-------- ( -30.90, z-score = -1.71, R) >droVir3.scaffold_12875 18240932 77 - 20611582 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAUUGAG------ (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))......------ ( -30.90, z-score = -1.87, R) >droWil1.scaffold_181089 8540221 79 + 12369635 GGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAGGAUUA----- ((((((((..((((.(((.(.((((....))))).))).))))......((((.......)))))))))))).......----- ( -32.80, z-score = -2.28, R) >droPer1.super_4 5395888 83 + 7162766 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAUUCUCAGAUUA (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))............ ( -30.90, z-score = -1.71, R) >dp4.chr3 10077553 83 + 19779522 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAUUUUCAGCUUA (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))............ ( -30.90, z-score = -1.70, R) >droAna3.scaffold_13266 12268387 81 + 19884421 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAUCGCUAAAC-- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))..........-- ( -30.90, z-score = -1.70, R) >droSim1.chr2R 19417110 75 + 19596830 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAGCA-------- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))....-------- ( -30.90, z-score = -1.74, R) >droSec1.super_23 822271 75 + 989336 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAGCA-------- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))....-------- ( -30.90, z-score = -1.74, R) >droEre2.scaffold_4845 22371283 75 + 22589142 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAGCA-------- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......))))))))))))....-------- ( -30.90, z-score = -1.74, R) >droYak2.chr2R 20933507 78 + 21139217 GGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAGCAGCA----- (((((((...((((.-..(..((((....))))..)...)))).....(((((.......)))))))))))).......----- ( -30.90, z-score = -1.33, R) >consensus GGGGACGUAGCUCAG_UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCCAGCA________ (((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))............ (-31.23 = -30.93 + -0.30)

| Location | 20,964,287 – 20,964,362 |
|---|---|
| Length | 75 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 89.53 |
| Shannon entropy | 0.23596 |
| G+C content | 0.60900 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
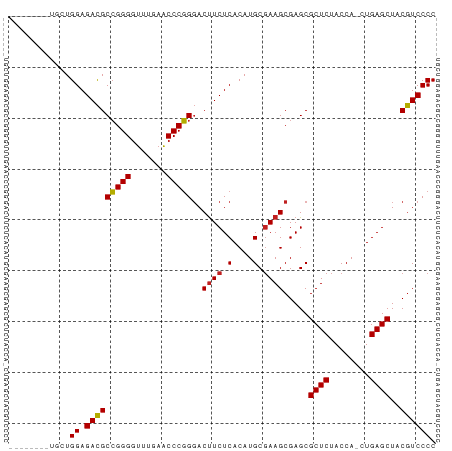
>dm3.chr2R 20964287 75 - 21146708 --------UGUUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC --------....((.(((((((((.......)))))(.((((.(....).)))))....((((.....-..))))..)))))). ( -25.20, z-score = -0.76, R) >droGri2.scaffold_15112 678172 81 + 5172618 --CGUAGCAAUUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC --((((((...(((((((((((((((....))))....((((.(....).)))))).))).))).)))-....))))))..... ( -27.80, z-score = -1.19, R) >droMoj3.scaffold_6496 22869371 75 - 26866924 --------CGCUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC --------(((((((((.(((.((((....)))).)).).)))).)).)))..(((...((((.....-..))))..))).... ( -26.90, z-score = -1.16, R) >droVir3.scaffold_12875 18240932 77 + 20611582 ------CUCAAUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC ------......((.(((((((((.......)))))(.((((.(....).)))))....((((.....-..))))..)))))). ( -25.20, z-score = -1.00, R) >droWil1.scaffold_181089 8540221 79 - 12369635 -----UAAUCCUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCC -----.......((((.(((((((.......))))))).)))).....(((.....)))((((........))))......... ( -23.70, z-score = -1.60, R) >droPer1.super_4 5395888 83 - 7162766 UAAUCUGAGAAUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC ............((.(((((((((.......)))))(.((((.(....).)))))....((((.....-..))))..)))))). ( -25.20, z-score = -0.79, R) >dp4.chr3 10077553 83 - 19779522 UAAGCUGAAAAUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC ...(((....(((((((.(((.((((....)))).)).).)))).)))....)))....((((.....-..))))......... ( -27.70, z-score = -1.64, R) >droAna3.scaffold_13266 12268387 81 - 19884421 --GUUUAGCGAUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC --((((.((.(((((((.(((.((((....)))).)).).)))).))))).))))....((((.....-..))))......... ( -28.10, z-score = -1.20, R) >droSim1.chr2R 19417110 75 - 19596830 --------UGCUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC --------....((.(((((((((.......)))))(.((((.(....).)))))....((((.....-..))))..)))))). ( -25.20, z-score = -0.72, R) >droSec1.super_23 822271 75 - 989336 --------UGCUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC --------....((.(((((((((.......)))))(.((((.(....).)))))....((((.....-..))))..)))))). ( -25.20, z-score = -0.72, R) >droEre2.scaffold_4845 22371283 75 - 22589142 --------UGCUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC --------....((.(((((((((.......)))))(.((((.(....).)))))....((((.....-..))))..)))))). ( -25.20, z-score = -0.72, R) >droYak2.chr2R 20933507 78 - 21139217 -----UGCUGCUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCC -----.(((((((((((.(((.((((....)))).)).).)))).)).))..)))....((((.....-..))))......... ( -26.30, z-score = -0.64, R) >consensus ________UGCUGGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA_CUGAGCUACGUCCCC ............((.(((((((((.......)))))..((((.(....).)))).....((((........))))..)))))). (-24.36 = -24.22 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:45 2011