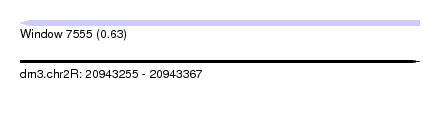
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,943,255 – 20,943,367 |
| Length | 112 |
| Max. P | 0.629668 |

| Location | 20,943,255 – 20,943,367 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.18 |
| Shannon entropy | 0.08182 |
| G+C content | 0.41830 |
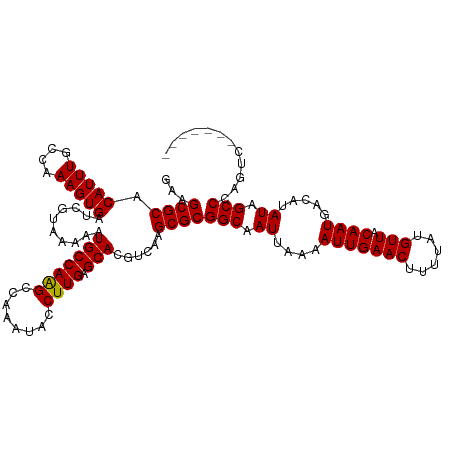
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.03 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.74 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
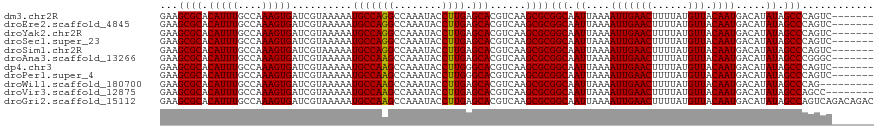
>dm3.chr2R 20943255 112 - 21146708 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAGGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -23.70, z-score = -0.60, R) >droEre2.scaffold_4845 22323029 112 - 22589142 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAGGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -23.70, z-score = -0.60, R) >droYak2.chr2R 20911023 112 - 21139217 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAGGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -23.70, z-score = -0.60, R) >droSec1.super_23 800917 112 - 989336 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAGGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -23.70, z-score = -0.60, R) >droSim1.chr2R 19395832 112 - 19596830 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAGGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -23.70, z-score = -0.60, R) >droAna3.scaffold_13266 12246247 112 - 19884421 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAAGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCGGGC------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -24.00, z-score = -0.15, R) >dp4.chr3 10054302 112 - 19779522 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAAGCCAAAUACCUUGGGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC------- ...((((.(((((....)))))..........(((((((........))).))))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -24.00, z-score = -0.39, R) >droPer1.super_4 5372670 112 - 7162766 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAAGCCAAAUACCUUGGGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC------- ...((((.(((((....)))))..........(((((((........))).))))......))))(((.((....(((((((......))).)))).....)).))).....------- ( -24.00, z-score = -0.39, R) >droWil1.scaffold_180700 5162051 110 + 6630534 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAAGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAG--------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).)))...--------- ( -24.00, z-score = -1.44, R) >droVir3.scaffold_12875 18209561 111 + 20611582 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAAGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCAGCC-------- ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).)))....-------- ( -24.90, z-score = -1.44, R) >droGri2.scaffold_15112 4525698 119 - 5172618 GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAAGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCAGUCAGACAGAC ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).))).(((.....))) ( -25.60, z-score = -1.29, R) >consensus GAAGCGCACAUUUGCCAAAGUGAUCGUAAAAAUGCCAAGCCAAAUACCUUGAGCACGUCAAGCGCGGCAAUUAAAAUUGAACUUUUAUGUUACAAUGACAUAUAGCCCAGUC_______ ...((((.(((((....)))))..........(((((((........)))).)))......))))(((.((....(((((((......))).)))).....)).)))............ (-24.27 = -24.03 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:41 2011