
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,899,003 – 20,899,130 |
| Length | 127 |
| Max. P | 0.870343 |

| Location | 20,899,003 – 20,899,130 |
|---|---|
| Length | 127 |
| Sequences | 5 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 73.51 |
| Shannon entropy | 0.46216 |
| G+C content | 0.46188 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.76 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.574883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
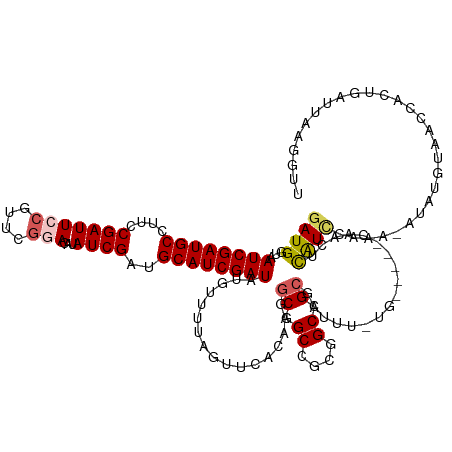
>dm3.chr2R 20899003 127 + 21146708 GAUGGUAAUCGAUGCCUUCCGAUUCCAUUCGGACAUAUCGAUGCAUCGAUAUGUCGUAGUUCACACGACGGCCGCGGCAGCGCUUUCUGA----AAACACAUACAUCGAUAUGUCAACACUGAUUAAAAUU (((((.(((((........))))))))))..((((((((((((.........(((((.(....))))))(((.((....)))))......----.........))))))))))))................ ( -36.80, z-score = -1.98, R) >droEre2.scaffold_4845 22274376 129 + 22589142 GAUGGUAAUCGAUGCCUUCCGAUUUCUUCCACA--UAUCGAUGCAUCGAUACGAUUUAGUUCGUCACGCGGCCGCAGCAGCGCUUUGUGCUUAUCGACACAUCCAUACAUAUUUUACCAAAGAUAGAGAUU ..(((((((((((((....((((..........--.))))..)))))))).((((..(((.((...(((.((....)).)))...)).))).))))..................)))))............ ( -29.80, z-score = -0.80, R) >droYak2.chr2R 20863975 128 + 21139217 GAUGGUAAUCGAUGCCUUCCGAUUACCUUCGCAC-UAUCGAUGCAUCGAUUCUAUUUAGUUCAUCACGCGGCCGCAGCAGCGCUUUGUGCUUACCGAGUUGUUAAGACACACAU--CCAAAGACAGAGGUU ((.((((((((........)))))))).))((((-....((((.((..(......)..)).)))).(((.((....)).)))....))))..(((...(((((...........--.....))))).))). ( -31.29, z-score = -0.33, R) >droSec1.super_23 756271 121 + 989336 GAUGGUAAUCGAUGCCUUCCGAUUCCGUUCGGACAUAUCGAUGCAUCGAUAUGUUUUAGUACACAAGGCGGCCACGGCAGCGCUUU--------GAGCUCAUCGAAU--UGUGAAGUCACUGAUUAAGGUU (((..((((((.((.((((((((((.((..(((((((((((....)))))))))))....)).(((((((((....))..))))))--------)........))))--)).)))).)).))))))..))) ( -39.90, z-score = -2.16, R) >droSim1.chr2R 19350907 121 + 19596830 GAUGGUAAUCGAUGCCUUCCGAUUCCGUUCGGACAUAUCGAUGCAUCGAUAUGUUUUAGUACACAAGGCGGCCACGGCAGCGCUUU--------GAGCUCAUCGAAU--UGUGAAGUCACUGAUUAAGGUU (((..((((((.((.((((((((((.((..(((((((((((....)))))))))))....)).(((((((((....))..))))))--------)........))))--)).)))).)).))))))..))) ( -39.90, z-score = -2.16, R) >consensus GAUGGUAAUCGAUGCCUUCCGAUUCCGUUCGGACAUAUCGAUGCAUCGAUAUGUUUUAGUUCACAAGGCGGCCGCGGCAGCGCUUU_UG_____CAACUCAUCCAAA_AUAUGUAACCACUGAUUAAGGUU ((((...((((((((....(((((((....)))...))))..)))))))).................((.((....)).))..................))))............................ (-18.20 = -18.76 + 0.56)

| Location | 20,899,003 – 20,899,130 |
|---|---|
| Length | 127 |
| Sequences | 5 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Shannon entropy | 0.46216 |
| G+C content | 0.46188 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.44 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
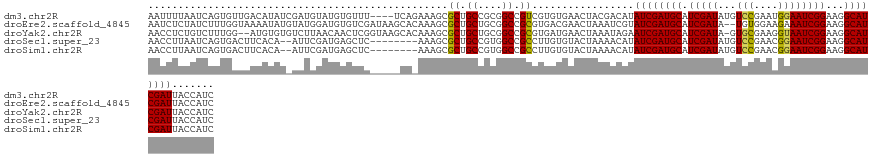
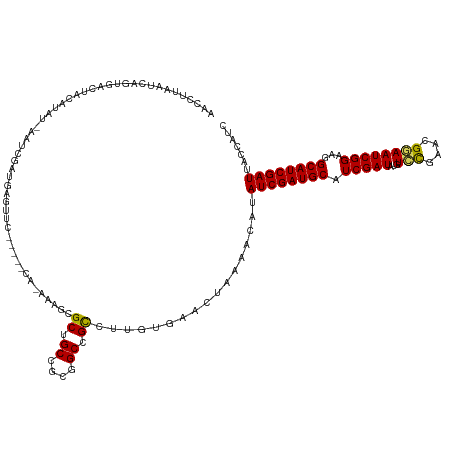
>dm3.chr2R 20899003 127 - 21146708 AAUUUUAAUCAGUGUUGACAUAUCGAUGUAUGUGUUU----UCAGAAAGCGCUGCCGCGGCCGUCGUGUGAACUACGACAUAUCGAUGCAUCGAUAUGUCCGAAUGGAAUCGGAAGGCAUCGAUUACCAUC .....(((((.(((((..(((((((((((((((((((----.....)))))).((....)).((((((....).)))))......)))))))))))))(((((......))))).))))).)))))..... ( -43.00, z-score = -2.48, R) >droEre2.scaffold_4845 22274376 129 - 22589142 AAUCUCUAUCUUUGGUAAAAUAUGUAUGGAUGUGUCGAUAAGCACAAAGCGCUGCUGCGGCCGCGUGACGAACUAAAUCGUAUCGAUGCAUCGAUA--UGUGGAAGAAAUCGGAAGGCAUCGAUUACCAUC ............((((((..(((((((.(((((((((((.........((((.((....)).)))).((((......))))))))))))))).)))--))))...((..((....))..))..)))))).. ( -40.30, z-score = -2.25, R) >droYak2.chr2R 20863975 128 - 21139217 AACCUCUGUCUUUGG--AUGUGUGUCUUAACAACUCGGUAAGCACAAAGCGCUGCUGCGGCCGCGUGAUGAACUAAAUAGAAUCGAUGCAUCGAUA-GUGCGAAGGUAAUCGGAAGGCAUCGAUUACCAUC ....(((((..((((--...((((.((((.(.....).))))))))..((((.((....)).))))......)))))))))((((((((.(((((.-.(((....))))))))...))))))))....... ( -36.20, z-score = -0.62, R) >droSec1.super_23 756271 121 - 989336 AACCUUAAUCAGUGACUUCACA--AUUCGAUGAGCUC--------AAAGCGCUGCCGUGGCCGCCUUGUGUACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGAUUACCAUC .....(((((.(((....))).--....((((.((.(--------((.(((..((....))))).))).)).((....((((((((....))))))))(((((......))))))).)))))))))..... ( -33.50, z-score = -1.58, R) >droSim1.chr2R 19350907 121 - 19596830 AACCUUAAUCAGUGACUUCACA--AUUCGAUGAGCUC--------AAAGCGCUGCCGUGGCCGCCUUGUGUACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGAUUACCAUC .....(((((.(((....))).--....((((.((.(--------((.(((..((....))))).))).)).((....((((((((....))))))))(((((......))))))).)))))))))..... ( -33.50, z-score = -1.58, R) >consensus AACCUUAAUCAGUGACUACAUAU_AAUCGAUGAGUUC_____CA_AAAGCGCUGCCGCGGCCGCCUUGUGAACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGAUUACCAUC ..................................................((.((....)).)).................((((((((.(((((...(((....))))))))...))))))))....... (-20.80 = -20.44 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:39 2011