| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,879,836 – 20,879,956 |
| Length | 120 |
| Max. P | 0.931324 |

| Location | 20,879,836 – 20,879,956 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Shannon entropy | 0.49005 |
| G+C content | 0.54940 |
| Mean single sequence MFE | -46.49 |
| Consensus MFE | -10.07 |
| Energy contribution | -9.36 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.83 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
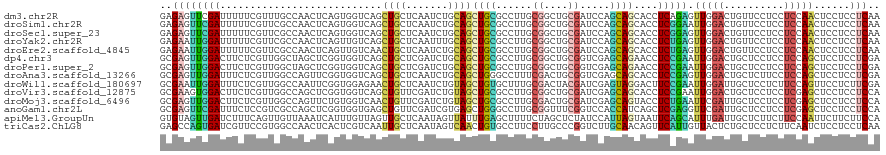
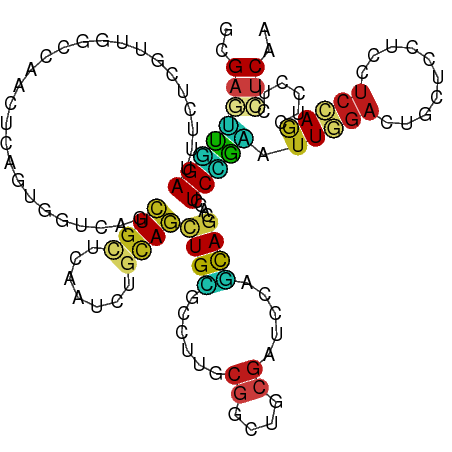
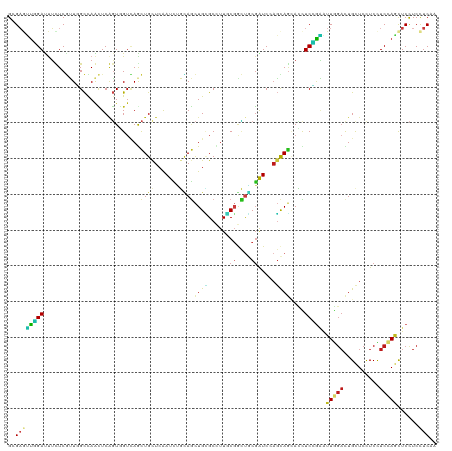
>dm3.chr2R 20879836 120 + 21146708 GAGAGUUCGAUUUUUCGUUUGCCAACUCAGUGGUCAGCUGCUCAAUCUGCAGCUGCGCCUUGCGGCUGCGAUCCAGCAGCACCUCAGAGUUGGACUGUUCCUCCUCCAACUCCUCCUCAA (((((((.((...........(((((((.(.(((..((((((..(((.((((((((.....)))))))))))..))))))))).).)))))))...........)).)))).)))..... ( -48.15, z-score = -4.25, R) >droSim1.chr2R 19331414 120 + 19596830 GAGAGUUCGAUUUUUCGUUCGCCAACUCAGUGGUCAGCUGCUCAAUCUGCAGCUGCGCCUUGCGGCUGCGAUCCAGCAGCACCUCGGAAUUGGACUGUUCCUCCUCCAACUCCUCCUCAA ..((((((((........)))..))))).(.(((..((((((..(((.((((((((.....)))))))))))..))))))))).)(((.(((((..........))))).)))....... ( -45.10, z-score = -3.53, R) >droSec1.super_23 736719 120 + 989336 GAGAGUUCGAUUUUUCGUUCGCCAACUCAGUGGUCAGCUGCUCAAUCUGCAGCUGCGCCUUGCGGCUGCGAUCCAGCAGCACCUCGGAGUUGGACUGUUCCUCCUCCAACUCCUCCUCAA ..((((((((........)))..))))).(.(((..((((((..(((.((((((((.....)))))))))))..))))))))).)(((((((((..........)))))))))....... ( -51.10, z-score = -4.87, R) >droYak2.chr2R 20844222 120 + 21139217 GAGAAUUGGAUUUUUCGUUCGCCAACUCAGUUGUCAGCUGCUCAAUUUGCAGCUGCGCCUUGCGGCUGCGAUCCAGCAGCACCUCUGAGUUGGACUGUUCCUCCUCCAACUCCUCCUCAA (((..(((((...........(((((((((..((..((((((....((((((((((.....))))))))))...))))))))..)))))))))...........)))))...)))..... ( -49.75, z-score = -5.48, R) >droEre2.scaffold_4845 22254656 120 + 22589142 GAGAAUUGGAUUUUUCGUUCGCCAACUCAGUUGUCAACUGCUCAAUCUGCAGCUGCGCCUUGCGGCUGCGAUCCAGCAGCACCUCUGAGUUGGACUGUUCCUCCUCCAACUCCUCCUCAA (((..(((((...........(((((((((..((...(((((..(((.((((((((.....)))))))))))..))))).))..)))))))))...........)))))...)))..... ( -46.75, z-score = -5.18, R) >dp4.chr3 1993204 120 + 19779522 GCGAGUUGGACUUCUCGUUGGCUAGCUCGGUGGUCAGCUGCUCGAUCUGCAGCUGCGCCUUGCGGCUGCGGUCGAGCAGAACCUCCGAAUUGGACUGCUCCUCCUCCAGCUCCUCCUCGA ..((((((((......((...((((.((((.(((...((((((((.((((((((((.....)))))))))))))))))).))).)))).))))...))......))))))))........ ( -60.50, z-score = -4.24, R) >droPer1.super_2 2166312 120 + 9036312 GCGAGUUGGACUUCUCGUUGGCUAGCUCGGUGGUCAGCUGCUCGAUCUGCAGCUGCGCCUUGCGGCUGCGGUCGAGCAGAACCUCCGAAUUGGACUGCUCCUCCUCCAGCUCCUCCUCGA ..((((((((......((...((((.((((.(((...((((((((.((((((((((.....)))))))))))))))))).))).)))).))))...))......))))))))........ ( -60.50, z-score = -4.24, R) >droAna3.scaffold_13266 12173031 120 + 19884421 GCGAGUUGGAUUUCUCGUUGGCCAGUUCGGUGGUCAGCUGCUCAAUCUGCAGCUGGGCCUUUCGACUGCGGUCGAGCAGCACCUCCGAGUUGGACUGCUCUUCCUCCAGCUCCUCCUCGA ..((((((((......((...(((..((((.(((..(((((((...((((((.(((.....))).))))))..)))))))))).))))..)))...))......))))))))........ ( -52.00, z-score = -2.45, R) >droWil1.scaffold_180697 1905060 120 + 4168966 GCGAAUUGGAUUUCUCGUUGGCCAAUUCGGUGGAGAACUGCUCAAUCUGUAGCUGUGCUUUGCGACUACGAUCGAGUAGGACUUCCGAAUUGGAUUGCUCCUCUUCCAGUUCCUCUUCGA ..((((((((......((...(((((((((..(((..((((((.(((.((((.(((.....))).))))))).))))))..))))))))))))...))......))))))))........ ( -44.70, z-score = -3.60, R) >droVir3.scaffold_12875 3450083 120 - 20611582 GCGAAGUGGACUUCUCGUUGGCCAGCUCGGUGGUCAGCUGUUCGAUCUGUAGCUGCGCCUUGCGGCUGCGAUCGAGCAGCACCUCCGAAUUGGACUGCUCCUCCUCGAGCUCCUCCUCCA (.((.(.(((.........((((((.((((.(((..(((((((((((.((((((((.....)))))))))))))))))))))).)))).)))).))((((......))))))).).))). ( -57.00, z-score = -3.92, R) >droMoj3.scaffold_6496 10840313 120 + 26866924 GCGAGUUGGACUUCUCGUUGGCCAGUUCUGUGGUCAACUGUUCGAUCUGUAGCUGCGCCUUGCGACUGCGAUCGAGCAGUACCUCUGAAUUCGAUUGCUCCUCCUCGAGCUCCUCCUCCA (.(((..(((....(((.((((((......))))))(((((((((((.((((.(((.....))).)))))))))))))))...........)))..((((......)))))))..)))). ( -43.40, z-score = -2.72, R) >anoGam1.chr2L 2178825 120 - 48795086 GCGAGUUCGAUUUCUCCGUCGCCAGCUCGGUGGUGAGCUGUUCGAUCGUGAGCUGGGCCUUGCGGUUUCGGUCCACCAUCAGCUCGAGGUUCGAUUGCUCCUCCUCGAGCUCCUCCUCCA ..(((((((((......))))..)))))(((((((..(((...(((((..((......))..))))).)))..)))))))(((((((((...((....))..)))))))))......... ( -48.10, z-score = -1.83, R) >apiMel3.GroupUn 309881929 120 - 399230636 GUGUAGUUGAUCUUUCAGUUGUUAAAUCAUUUGUUAGUUGCUCAAUAGUUAUUUGAGCUUUUCUAGCUCUAUCCAUUAGUAAUUCAGCAUUUGAUUGCUCUUCUUCCAAUUCUUCUUCCA ...((..(((....)))..))...(((((..(((((((((((.(((.(......(((((.....)))))...).)))))))))).))))..)))))........................ ( -20.10, z-score = -1.76, R) >triCas2.ChLG8 12167536 120 + 15773733 GAGCCAGUGAUCGUUCCGUGGCCAACUCACUCGUCAAUUGCUCAAUAGUCAACUGUGCCUUCCUUGCCCGGUCUUGCAACAGUUCAUUGUUACUCUGCUCCUCUUCAAUCUCCUCCUCAA ((((...(((.((....((((.....)))).)))))...))))((((((.((((((((...((......))....)).)))))).))))))............................. ( -23.70, z-score = -1.67, R) >consensus GCGAGUUGGAUUUCUCGUUGGCCAACUCAGUGGUCAGCUGCUCAAUCUGCAGCUGCGCCUUGCGGCUGCGAUCCAGCAGCACCUCCGAAUUGGACUGCUCCUCCUCCAGCUCCUCCUCAA ..((((((((...........................((((.......))))((((......((....)).....))))....))))).(((((..........)))))......))).. (-10.07 = -9.36 + -0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:36 2011