| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,847,448 – 20,847,577 |
| Length | 129 |
| Max. P | 0.999458 |

| Location | 20,847,448 – 20,847,577 |
|---|---|
| Length | 129 |
| Sequences | 8 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 63.33 |
| Shannon entropy | 0.72813 |
| G+C content | 0.57014 |
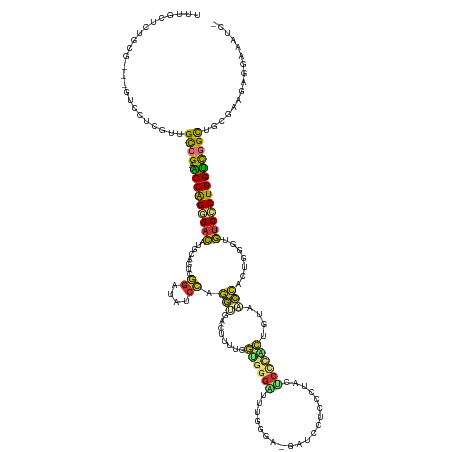
| Mean single sequence MFE | -56.61 |
| Consensus MFE | -22.74 |
| Energy contribution | -21.52 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.79 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
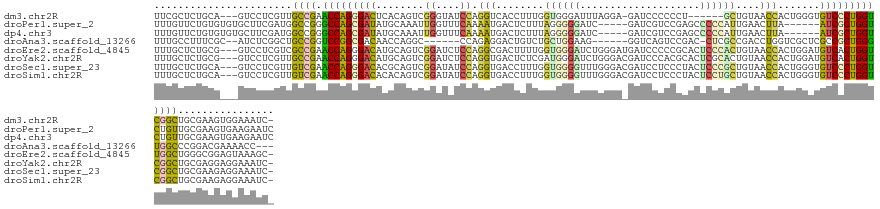
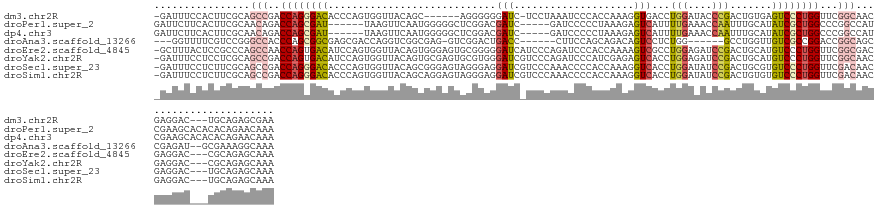
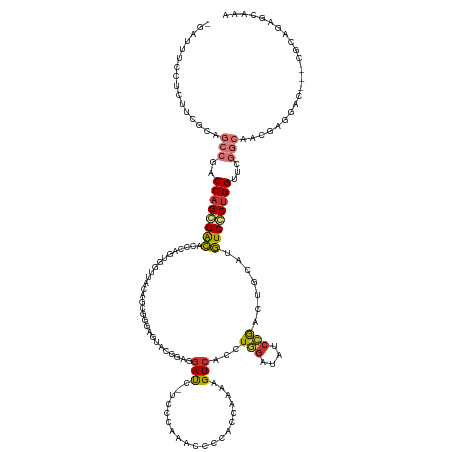
>dm3.chr2R 20847448 129 + 21146708 UUCGCUCUGCA---GUCCUCGUUGCCGAACCAGGGACUCACAGUCGGGUAUCCAGGUCACCUUUGGUGGGAUUUAGGA-GAUCCCCCCU------GCUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAAGUGGAAAUC- ((((((.((((---(((.(((....)))(((((((((..(((((.(((...(((((.....))))).((((((.....-))))))))).------))))).(((....)))))))))))).))))))).))))))....- ( -57.00, z-score = -2.84, R) >droPer1.super_2 2125451 129 + 9036312 UUUGUUCUGUGUGUGCUUCGAUGGCCGGGCCAGCGAUAUGCAAAUUGGUUUCAAAAUGACUCUUUAGGGGGAUC-----GAUCGUCCGAGCCCCCAUUGAACUUA------AUCGCUGGUCUGUUGCGAAGUGAAGAAUC ...(((((.....(((((((.....((((((((((((.....((..((((.......))))..)).(((((.((-----(......))).)))))..........------))))))))))))...))))))).))))). ( -46.80, z-score = -3.00, R) >dp4.chr3 1952411 129 + 19779522 UUUGUUCUGUGUGUGCUUCGAUGGCCGGGCCAGCGAUAUGCAAAUUGGUUUCAAAAUGACUCUUUAGGGGGAUC-----GAUCGUCCGAGCCCCCAUUGAACUUA------AUCGCUGGUCUGUUGCGAAGUGAAGAAUC ...(((((.....(((((((.....((((((((((((.....((..((((.......))))..)).(((((.((-----(......))).)))))..........------))))))))))))...))))))).))))). ( -46.80, z-score = -3.00, R) >droAna3.scaffold_13266 12138300 122 + 19884421 UUUGCCUUUCGC--AUCUCGGCUGCCGGUCCGGCGACAACCAGGC------CCAGAGGACUGUCUGCUGGAAG------GGUCAGUCCGAC-CUCGCCGACCUGGUCGCUCGCCGCUGGGUGGCCCGGACGAAAACC--- ......(((((.--.(((.(((..((.(..(((((((.((((((.------.((((......)))).(((..(------((((.....)))-))..))).)))))).).)))))).).))..))).))))))))...--- ( -53.10, z-score = -0.66, R) >droEre2.scaffold_4845 22220433 136 + 22589142 UUUGCUCUGCG---GUCCUCGUCGCCGAACCAGGGACAUGCAGUCGGAUCUCCAGGCGACUUUUGGUGGGAUCUGGGAUGAUCCCCCGCACUCCCACUGUAACCACUGGAUGUCACUGGUUGGCUGGGCGGAGUAAAGC- (((((((((((---(......))(((.((((((.(((((.((((.((....)).((..((....(((((((..((((.......))))...)))))))))..)))))).))))).)))))))))...))))))))))..- ( -59.40, z-score = -2.24, R) >droYak2.chr2R 20811328 136 + 21139217 UUUGCUCUGCG---GUCCUCGUUGCCGAACCAGGGACAUGCAGUCGGAUCUCCAGGUGACUCUCGAUGGGAUCUGGGACGAUCCCACGCACUCGCACUGUAACCACUGGAUGUCACUGGUCGGCUGCGAGGAGGAAAUC- ...........---.(((((((.(((((.((((.(((((.((((.((.....(((((((....((.(((((((......)))))))))...)))).)))...)))))).))))).))))))))).))))))).......- ( -63.00, z-score = -3.70, R) >droSec1.super_23 704419 136 + 989336 UUUGCUCUGCA---GUCCUCGUUGUCGAACCAGGGACACGCAGUCGGAUAUCCAGGUGACCUUUGGUGGGGUUUGGGACGAUCCUCCCUACUCCCGCUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAAGAGGAAAUC- (((.(((((((---(((.(((....)))(((((((((((.((((.((....)).(((.((....((((((((..((((......)))).)).)))))))).))))))).))))))))))).))))))..)))).)))..- ( -65.60, z-score = -4.22, R) >droSim1.chr2R 19299248 136 + 19596830 UUUGCUCUGCA---GUCCUCGUUGUCGAACCAGGGACACACAGUCGGAUAUCCAGGUGACCUUUGGUGGGGUUUGGGACGAUCCUCCCUACUCCUGCUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAAGAGGAAAUC- (((.(((((((---(((.(((....)))(((((((((((.((((.((.(((.((((........(((((((...(((.....))))))))))))))..))).)))))).))))))))))).))))))..)))).)))..- ( -61.20, z-score = -3.33, R) >consensus UUUGCUCUGCG___GUCCUCGUUGCCGAACCAGGGACAUGCAGUCGGAUAUCCAGGUGACUUUUGGUGGGAUUUGGGA_GAUCCUCCCUACUCCCACUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAAGAGGAAAUC_ .......................((((.(((((((((........((....)).(((........((((((....................))))))....))).......)))))))))))))................ (-22.74 = -21.52 + -1.22)
| Location | 20,847,448 – 20,847,577 |
|---|---|
| Length | 129 |
| Sequences | 8 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 63.33 |
| Shannon entropy | 0.72813 |
| G+C content | 0.57014 |
| Mean single sequence MFE | -50.50 |
| Consensus MFE | -13.11 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20847448 129 - 21146708 -GAUUUCCACUUCGCAGCCGACCAGGGACACCCAGUGGUUACAGC------AGGGGGGAUC-UCCUAAAUCCCACCAAAGGUGACCUGGAUACCCGACUGUGAGUCCCUGGUUCGGCAACGAGGAC---UGCAGAGCGAA -......(.((..((((((.(((((((((.(.((((((((((...------..(((((((.-......))))).))....))))))(((....))))))).).)))))))))(((....))))).)---)))..)).).. ( -57.80, z-score = -3.73, R) >droPer1.super_2 2125451 129 - 9036312 GAUUCUUCACUUCGCAACAGACCAGCGAU------UAAGUUCAAUGGGGGCUCGGACGAUC-----GAUCCCCCUAAAGAGUCAUUUUGAAACCAAUUUGCAUAUCGCUGGCCCGGCCAUCGAAGCACACACAGAACAAA .........(((((...(.(.((((((((------..........(((((.(((......)-----)).)))))......(.((..(((....)))..)))..)))))))).).).....)))))............... ( -33.50, z-score = -1.32, R) >dp4.chr3 1952411 129 - 19779522 GAUUCUUCACUUCGCAACAGACCAGCGAU------UAAGUUCAAUGGGGGCUCGGACGAUC-----GAUCCCCCUAAAGAGUCAUUUUGAAACCAAUUUGCAUAUCGCUGGCCCGGCCAUCGAAGCACACACAGAACAAA .........(((((...(.(.((((((((------..........(((((.(((......)-----)).)))))......(.((..(((....)))..)))..)))))))).).).....)))))............... ( -33.50, z-score = -1.32, R) >droAna3.scaffold_13266 12138300 122 - 19884421 ---GGUUUUCGUCCGGGCCACCCAGCGGCGAGCGACCAGGUCGGCGAG-GUCGGACUGACC------CUUCCAGCAGACAGUCCUCUGG------GCCUGGUUGUCGCCGGACCGGCAGCCGAGAU--GCGAAAGGCAAA ---((((.(((((((((...)))...(((((.((((((((((((...(-(((.....))))------...))..((((......)))))------))))))))))))))))).))).))))....(--((.....))).. ( -55.10, z-score = -0.81, R) >droEre2.scaffold_4845 22220433 136 - 22589142 -GCUUUACUCCGCCCAGCCAACCAGUGACAUCCAGUGGUUACAGUGGGAGUGCGGGGGAUCAUCCCAGAUCCCACCAAAAGUCGCCUGGAGAUCCGACUGCAUGUCCCUGGUUCGGCGACGAGGAC---CGCAGAGCAAA -(((((.((((.....(((((((((.(((((.((((((...(((.(.((.(..((((((((......)))))).))...).)).)))).....)).)))).))))).)))))).))).....))).---.).)))))... ( -56.10, z-score = -2.70, R) >droYak2.chr2R 20811328 136 - 21139217 -GAUUUCCUCCUCGCAGCCGACCAGUGACAUCCAGUGGUUACAGUGCGAGUGCGUGGGAUCGUCCCAGAUCCCAUCGAGAGUCACCUGGAGAUCCGACUGCAUGUCCCUGGUUCGGCAACGAGGAC---CGCAGAGCAAA -.......((((((..(((((((((.(((((.((((((...(((.(.((.(.(((((((((......))))))).))..).)).)))).....)).)))).))))).)))).)))))..)))))).---........... ( -59.20, z-score = -3.85, R) >droSec1.super_23 704419 136 - 989336 -GAUUUCCUCUUCGCAGCCGACCAGGGACACCCAGUGGUUACAGCGGGAGUAGGGAGGAUCGUCCCAAACCCCACCAAAGGUCACCUGGAUAUCCGACUGCGUGUCCCUGGUUCGACAACGAGGAC---UGCAGAGCAAA -......((((..((((((.(((((((((((.(((((((.((.(.(((.((.((((......))))..))))).).....)).)))(((....))))))).)))))))))))(((....))))).)---))))))).... ( -54.90, z-score = -2.61, R) >droSim1.chr2R 19299248 136 - 19596830 -GAUUUCCUCUUCGCAGCCGACCAGGGACACCCAGUGGUUACAGCAGGAGUAGGGAGGAUCGUCCCAAACCCCACCAAAGGUCACCUGGAUAUCCGACUGUGUGUCCCUGGUUCGACAACGAGGAC---UGCAGAGCAAA -......((((..((((((.(((((((((((.((((((......((((....((((......))))..(((........)))..)))).....)).)))).)))))))))))(((....))))).)---))))))).... ( -53.90, z-score = -2.70, R) >consensus _GAUUUCCUCUUCGCAGCCGACCAGCGACACCCAGUGGUUACAGCGGGAGUACGGAGGAUC_UCCCAAACCCCACCAAAAGUCACCUGGAUAUCCGACUGCAUGUCCCUGGUUCGGCAACGAGGAC___CGCAGAGCAAA ................(((..((((((((............................(((....................)))...(((....))).......))))))))...)))....................... (-13.11 = -13.24 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:33 2011