
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,846,676 – 20,846,778 |
| Length | 102 |
| Max. P | 0.619005 |

| Location | 20,846,676 – 20,846,778 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.13 |
| Shannon entropy | 0.62546 |
| G+C content | 0.47396 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.36 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
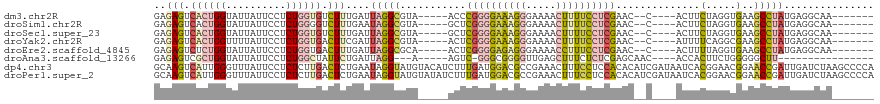
>dm3.chr2R 20846676 102 - 21146708 GAGAGUCACUGGUAUUAUUCCUCUGGUGUCUUUGAUUAGGCGUA-----ACCCGGGGAAAGGGAAAACUUUUCCUCGAAC--C----ACUUCUAGGUGAAGCCUAUGAGGCAA------- .((((.(((..(..........)..))).))))...(((((...-----...((((((((((.....))))))))))...--(----(((....))))..)))))........------- ( -33.70, z-score = -1.60, R) >droSim1.chr2R 19298453 102 - 19596830 GAGAGUCACUGGUAUUAUUCCUCUGGGGUCUUUGAAUAGGCGUA-----GCUCGGGGAAAGGGAAAACUUUUCCUCGAAC--C----ACUUCUAGGUGAAGCCUAUGAGGCAA------- ...................((((..(((.(((((..((((.((.-----(.(((((((((((.....))))))))))).)--.----)).))))..))))))))..))))...------- ( -34.30, z-score = -1.39, R) >droSec1.super_23 703630 102 - 989336 GAGAGUCACUGGUAUUAUUCCUCUGGUGUCUUUGAUUAGGCGUA-----GCUCGGGGAAAGGGAAAACUUUUCCUCGAAC--C----ACUUCUAGGUGAAGCCUAUGAGGCAA------- .((((.(((..(..........)..))).))))...(((((...-----..(((((((((((.....)))))))))))..--(----(((....))))..)))))........------- ( -35.10, z-score = -1.94, R) >droYak2.chr2R 20810523 102 - 21139217 GAGAGUCACUGGUUUUAUUCCUCUGGUGACUUCGAUUAGGCGUA-----ACUCGGGGAAAGGGAAAACUUUUCCUCGAAC--C----AUUUUCAGGCGAAGCCUAUGAGGCAA------- ..(((((((..(..........)..)))))))(...(((((...-----..(((((((((((.....))))))))))).(--(----.......))....)))))...)....------- ( -34.90, z-score = -2.19, R) >droEre2.scaffold_4845 22219615 102 - 22589142 GAGAGUCUCUGGUAUUAUUCCUCUGGUGACUUUGAUUAGGCGCA-----ACUCGGGGAGAGGGAAAACCUUUCCUCGAAC--C----ACUUUUAGGUGAAGCCUAUGAGGCAA------- .((((((.(..(..........)..).))))))...(((((...-----..(((((((((((.....)))))))))))..--(----(((....))))..)))))........------- ( -37.80, z-score = -2.51, R) >droAna3.scaffold_13266 12136653 91 - 19884421 GAGAGUCGCUGGUAUUAUUCCUCUGGCUAUUCUGAUUAGG---A-----AGUC-GGGCGGGGUUGAGCUUUCUCUCGAGCAAC----ACCACUUCUGGGGGCUU---------------- (((((..(((.(....((((((((((((.((((....)))---)-----))))-))).)))))).)))...)))))((((...----.(((....)))..))))---------------- ( -28.90, z-score = -0.22, R) >dp4.chr3 1951464 120 - 19779522 GCAAGUCAUUGGGUUUAUUCCUCUCUUGACUCUGAAUAGGUAUGUACAUCUUUGAUGGACGCCGAAACUUUCCUCCACACAUCGAUAAUCACGGAACGGAACCGAUUGAUCUAAGCCCCA ..........(((((((....((..(((..((((....(((..((.((((...)))).)))))......((((...................))))))))..)))..))..))))))).. ( -20.11, z-score = 1.78, R) >droPer1.super_2 2124508 120 - 9036312 GCAAGUCAUUGGGUUUAUUCCUCUCUUGACUCUGAAUAGGUAUGUAUAUCUUUGAUGGACGCCGAAACUUUCCUCCACACAUCGAUAAUCACGGAACGGAACCGAUUGAUCUAAGCCCCA ((..((((.(.(((((.((((................(((((....)))))((((((......................)))))).......))))..))))).).))))....)).... ( -18.75, z-score = 2.18, R) >consensus GAGAGUCACUGGUAUUAUUCCUCUGGUGACUUUGAUUAGGCGUA_____ACUCGGGGAAAGGGAAAACUUUUCCUCGAAC__C____ACUUCUAGGUGAAGCCUAUGAGGCAA_______ ..(((.((((((..........)))))).)))....(((((...........((((((((((.....))))))))))..............(.....)..)))))............... (-15.61 = -16.36 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:31 2011