
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,831,475 – 20,831,581 |
| Length | 106 |
| Max. P | 0.669925 |

| Location | 20,831,475 – 20,831,581 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 62.73 |
| Shannon entropy | 0.71214 |
| G+C content | 0.51654 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -11.15 |
| Energy contribution | -12.67 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.669925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20831475 106 + 21146708 CAUAAACGCAUACGUAUGUAUGUACACUUCCUAAGCGAUAAGGUGAGGAGCAAGAAUCAUACUCACCCAAGGCAGGGGUCCAGCAGACGGGGAGCCCUGUGGCGGC .......((((((....))))))...........((.....((((((..............))))))....((((((.(((.(....)..))).)))))).))... ( -31.94, z-score = -0.73, R) >droSim1.chr2R 19284288 102 + 19596830 ----CAUGAACAUGUACGUAUGUACAGUUCCCAAGCGUUAAGGUGAGGAGCAAGAAUCAUACUCACCCAAGGCAGGGGUCCAGCAGACGGGCAGCCCUGUGGCGGC ----...((((.(((((....))))))))).....((((..((((((..............))))))....((((((((((.......))))..)))))))))).. ( -33.24, z-score = -1.05, R) >droSec1.super_23 688544 102 + 989336 ----CAUGAACAUGUACGUAUGUACAGUUCCCAAGCGUUAAGGUGAGGAGCAAGAAUCAUACUCACCCAAGGCAGGGGUCCAGCAGACGGGCAGCCCUGUGGCGGC ----...((((.(((((....))))))))).....((((..((((((..............))))))....((((((((((.......))))..)))))))))).. ( -33.24, z-score = -1.05, R) >droYak2.chr2R 20795179 90 + 21139217 ----------------CAUAUGUACAUUUCAGAAGCGAUAAGGUGAGGAGUAAGAAUCAAACUCACCCCAGGCAGGGAUCCAGCAGUCGGGCCGCCCUGUGGCGGC ----------------...................((((..((((((..............))))))....((.((...)).)).)))).((((((....)))))) ( -24.84, z-score = 0.03, R) >droEre2.scaffold_4845 22204295 102 + 22589142 ----CAUACAUACAUAUGUAUGUACAUUUCAGAAGCGAUAAGGUGAGGAGCAAGAAUCAUACUCACCCAAGGCAGGGGUCCAGCAGUCGGGCAGCCCUGUGGCGGC ----..(((((((....)))))))..........((.....((((((..............))))))....((((((((((.......))))..)))))).))... ( -31.74, z-score = -1.73, R) >droMoj3.scaffold_6496 10773154 88 + 26866924 ------------------GUUAAACACUCAAAUAAUAUUAUUUUGAUAAUAAAACAUUUGAAUUACCCGCCGCACAGAUCCAAAAGGCGGGGUCCACGACGACGAC ------------------.........((((((..((((((....))))))....))))))....((((((..............))))))(((......)))... ( -18.64, z-score = -2.39, R) >droVir3.scaffold_12875 3378756 80 - 20611582 --------------------UAAAAGCUAAGGUAUACUCAUUACAGCAAUAA------UAACUUACCUGCCGCACAGCUCCAGCAGUCGGGGUAAGCGUCGGCGAC --------------------.....(((...(((.......)))........------...((((((..(((.((.((....)).)))))))))))....)))... ( -16.50, z-score = 0.27, R) >consensus _____A___A_A__UACGUAUGUACACUUCAGAAGCGUUAAGGUGAGGAGCAAGAAUCAUACUCACCCAAGGCAGGGGUCCAGCAGACGGGCAGCCCUGUGGCGGC ..................................((.....((((((..............))))))....((((((.(((.......)))...)))))).))... (-11.15 = -12.67 + 1.52)
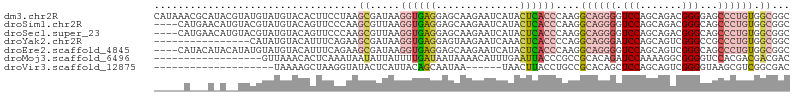

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:30 2011