
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,802,193 – 20,802,505 |
| Length | 312 |
| Max. P | 0.997646 |

| Location | 20,802,193 – 20,802,313 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.24 |
| Shannon entropy | 0.28851 |
| G+C content | 0.42481 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -17.10 |
| Energy contribution | -16.51 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.560495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
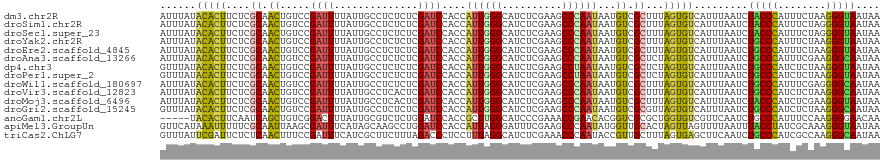
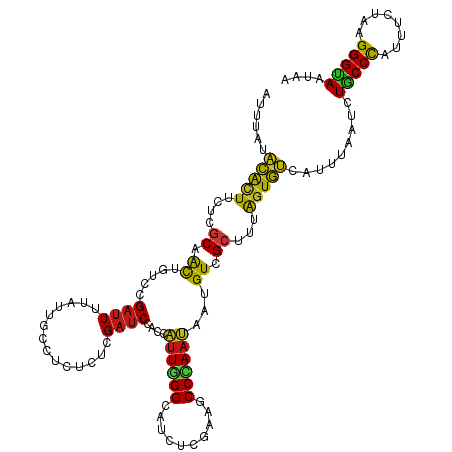
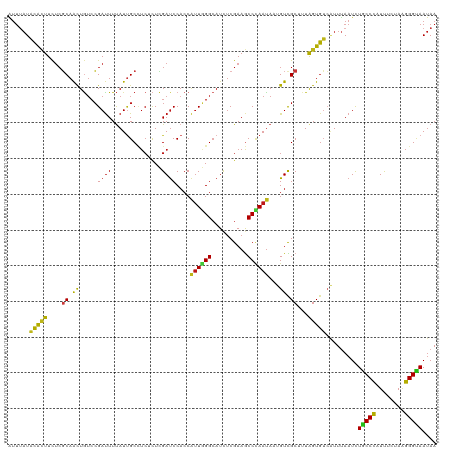
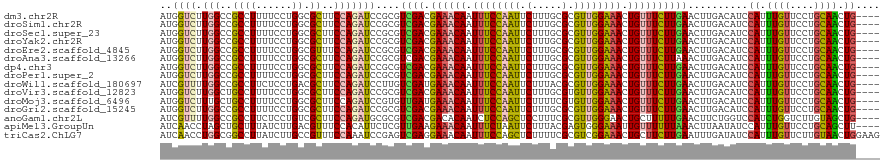
>dm3.chr2R 20802193 120 + 21146708 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAAGGGUAAUAA ......(((((....((.((....(((..............))).......(((((((........)))))))...)).))...))))).........(((((........))))).... ( -23.34, z-score = -1.99, R) >droSim1.chr2R 19254809 120 + 19596830 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAGGGGUAAUAA .............(((........)))..(((((((((((...........(((((((........)))))))((((.(((....))).))))................))))))))))) ( -26.80, z-score = -2.64, R) >droSec1.super_23 659197 120 + 989336 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAGGGGUAAUAA .............(((........)))..(((((((((((...........(((((((........)))))))((((.(((....))).))))................))))))))))) ( -26.80, z-score = -2.64, R) >droYak2.chr2R 20765650 120 + 21139217 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAA ......(((((....((.((....(((..............))).......(((((((........)))))))...)).))...))))).........(((((........))))).... ( -23.34, z-score = -1.58, R) >droEre2.scaffold_4845 22172265 120 + 22589142 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAA ......(((((....((.((....(((..............))).......(((((((........)))))))...)).))...))))).........(((((........))))).... ( -23.34, z-score = -1.58, R) >droAna3.scaffold_13266 12090605 120 + 19884421 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUUUCGAAGGGCAAUAA .............(((........)))..((((((((.((.((((......(((((((........)))))))((((.(((....))).)))).............)))))))))))))) ( -29.50, z-score = -3.00, R) >dp4.chr3 1892086 120 + 19779522 GUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAA .............(((........)))..(((((((((.....(((.....(((((((........)))))))((((.(((....))).))))..........))).....))))))))) ( -21.30, z-score = -0.88, R) >droPer1.super_2 2074867 120 + 9036312 GUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAA .............(((........)))..(((((((((.....(((.....(((((((........)))))))((((.(((....))).))))..........))).....))))))))) ( -21.30, z-score = -0.88, R) >droWil1.scaffold_180697 1832620 120 + 4168966 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCGAGGGGCAAUAA .............(((........)))..((((((((((((..........(((((((........)))))))((((.(((....))).))))...............)))))))))))) ( -32.90, z-score = -3.60, R) >droVir3.scaffold_12823 2414423 120 + 2474545 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCACUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAA .............(((........)))..(((((((((.....(((.....(((((((........)))))))((((.(((....))).))))..........))).....))))))))) ( -25.60, z-score = -2.12, R) >droMoj3.scaffold_6496 1760776 120 + 26866924 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCACUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUCUCGAAGGGUAAUAA .............(((........)))..(((((((((...((((......(((((((........)))))))((((.(((....))).)))).............)))).))))))))) ( -26.70, z-score = -2.56, R) >droGri2.scaffold_15245 2652053 120 - 18325388 GUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCGUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAA ......(((((...(((.((....(((..............))).......(((((((........)))))))...)).)))..))))).........(((((........))))).... ( -28.84, z-score = -2.95, R) >anoGam1.chr2L 41834918 115 + 48795086 -----UACACUUCAAUCAGCUGUCGGACUUUAUUGCGUCUCUGGAUCCACCGCUUGGCAUCCCGAAACCGAACACGGUCGCGCUGGUGUCGUUCAAUCUGCCCAUUUCCAAGGGGAACAA -----.........((((((.((..((((...(((.((.((.(((((((.....))).)))).)).)))))....))))))))))))...((((......(((........))))))).. ( -27.10, z-score = 1.04, R) >apiMel3.GroupUn 34969727 120 + 399230636 GUUCAUAAAUUUUUCGCAAUUAAGCGAUUUCAUAGCAAGCCUGGAUCCACCAUUAGGGAUUUCGAAGCCCAAUAUGGUUGCACUAGUUAGUUUUAAUUUACCUAUCGCAAAGGGUAAUAA .............((((......))))......(((.(((.(((..(.(((((..(((.........)))...))))).)..)))))).))).....((((((........))))))... ( -20.80, z-score = 0.38, R) >triCas2.ChLG7 4487131 120 - 17478683 GUUUAUUCGAUUCUCUCAACUUUCCGAUUUCAUCGCUUCUUUAGACCCUCCUUUAGGCAUCUCGAAACCCAAUACCGUUGCUUUAGUGAGCUUCAAUCUGCCCAUCGCCAAGGGCAAUAA ........((((............((((...))))........((..(((((..(((((...((...........)).))))).)).)))..))))))(((((........))))).... ( -19.60, z-score = -0.33, R) >consensus AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAA ......(((((....((.((.....((((..............))))....((((((..........))))))...)).))...))))).........(((((........))))).... (-17.10 = -16.51 + -0.58)
| Location | 20,802,193 – 20,802,313 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.24 |
| Shannon entropy | 0.28851 |
| G+C content | 0.42481 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -27.14 |
| Energy contribution | -27.73 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
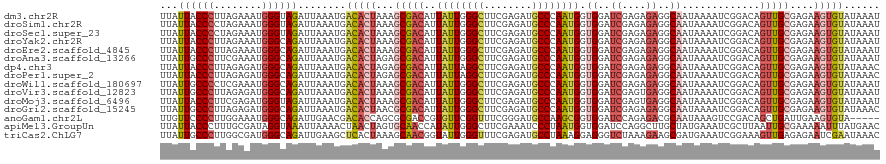
>dm3.chr2R 20802193 120 - 21146708 UUAUUACCCUUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -36.30, z-score = -4.22, R) >droSim1.chr2R 19254809 120 - 19596830 UUAUUACCCCUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -35.10, z-score = -3.65, R) >droSec1.super_23 659197 120 - 989336 UUAUUACCCCUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -35.10, z-score = -3.65, R) >droYak2.chr2R 20765650 120 - 21139217 UUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ......(((........)))...........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -32.50, z-score = -2.87, R) >droEre2.scaffold_4845 22172265 120 - 22589142 UUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ......(((........)))...........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -32.50, z-score = -2.87, R) >droAna3.scaffold_13266 12090605 120 - 19884421 UUAUUGCCCUUCGAAAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -38.30, z-score = -3.76, R) >dp4.chr3 1892086 120 - 19779522 UUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAC ......(((........)))...........(((((...(((((((((((.(((........))).))))))(..((....))..)..............)))))....)))))...... ( -26.00, z-score = -0.99, R) >droPer1.super_2 2074867 120 - 9036312 UUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAC ......(((........)))...........(((((...(((((((((((.(((........))).))))))(..((....))..)..............)))))....)))))...... ( -26.00, z-score = -0.99, R) >droWil1.scaffold_180697 1832620 120 - 4168966 UUAUUGCCCCUCGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -37.10, z-score = -3.34, R) >droVir3.scaffold_12823 2414423 120 - 2474545 UUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGUGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))...((((..............))))...)))))....)))))...... ( -37.24, z-score = -3.62, R) >droMoj3.scaffold_6496 1760776 120 - 26866924 UUAUUACCCUUCGAGAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGUGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))...((((..............))))...)))))....)))))...... ( -35.24, z-score = -3.08, R) >droGri2.scaffold_15245 2652053 120 + 18325388 UUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAACGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAC ...((((((........))))))........(((((..((((((((((((((((........))))))))))(..((....))..)..............))))))...)))))...... ( -40.00, z-score = -4.73, R) >anoGam1.chr2L 41834918 115 - 48795086 UUGUUCCCCUUGGAAAUGGGCAGAUUGAACGACACCAGCGCGACCGUGUUCGGUUUCGGGAUGCCAAGCGGUGGAUCCAGAGACGCAAUAAAGUCCGACAGCUGAUUGAAGUGUA----- ...((((....)))).(((((..((((..(((.((((((((....))))).))).)))((((.(((.....))))))).......))))...)))))(((.((......))))).----- ( -31.20, z-score = 0.81, R) >apiMel3.GroupUn 34969727 120 - 399230636 UUAUUACCCUUUGCGAUAGGUAAAUUAAAACUAACUAGUGCAACCAUAUUGGGCUUCGAAAUCCCUAAUGGUGGAUCCAGGCUUGCUAUGAAAUCGCUUAAUUGCGAAAAAUUUAUGAAC ..........(((((.(((...............))).))))).((((..((((((.((..((((.....).))))).))))))..))))...((((......))))............. ( -22.16, z-score = -0.17, R) >triCas2.ChLG7 4487131 120 + 17478683 UUAUUGCCCUUGGCGAUGGGCAGAUUGAAGCUCACUAAAGCAACGGUAUUGGGUUUCGAGAUGCCUAAAGGAGGGUCUAAAGAAGCGAUGAAAUCGGAAAGUUGAGAGAAUCGAAUAAAC (((((((((((..(..((((((..((((((((((.((..(...)..)).))))))))))..))))))..))))))).........((((....((((....))))....))))))))).. ( -33.70, z-score = -2.23, R) >consensus UUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((..((((((((........)))))))).((..((....))..)).............)))))....)))))...... (-27.14 = -27.73 + 0.59)
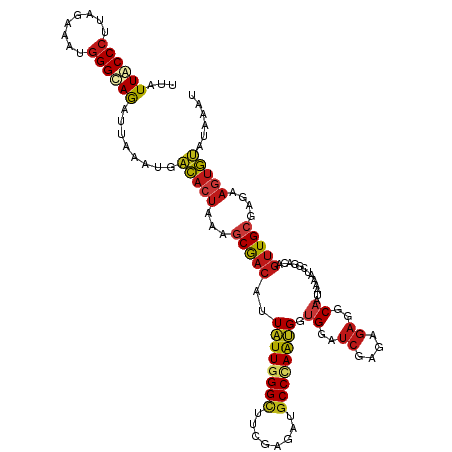
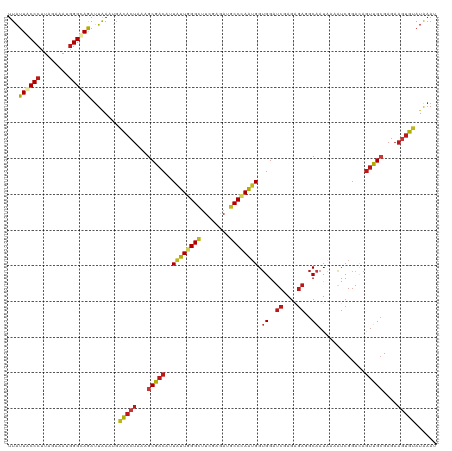
| Location | 20,802,233 – 20,802,353 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Shannon entropy | 0.26500 |
| G+C content | 0.45167 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -22.26 |
| Energy contribution | -21.51 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
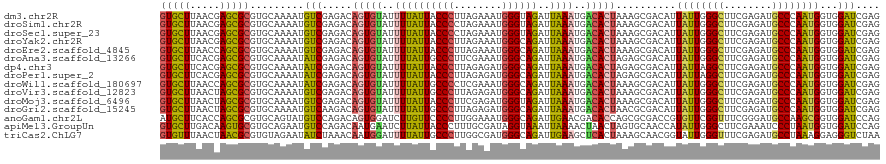
>dm3.chr2R 20802233 120 + 21146708 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -34.00, z-score = -4.38, R) >droSim1.chr2R 19254849 120 + 19596830 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAGGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -33.90, z-score = -3.87, R) >droSec1.super_23 659237 120 + 989336 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAGGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -33.90, z-score = -3.87, R) >droYak2.chr2R 20765690 120 + 21139217 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -34.00, z-score = -3.83, R) >droEre2.scaffold_4845 22172305 120 + 22589142 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUGGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -35.20, z-score = -3.63, R) >droAna3.scaffold_13266 12090645 120 + 19884421 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUUUCGAAGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...))))))).(((.(((....)))..))). ( -35.00, z-score = -2.98, R) >dp4.chr3 1892126 120 + 19779522 CUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCAC ......(((....)))((.((.(((.((.....(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))......))..))).)).)).. ( -30.80, z-score = -2.38, R) >droPer1.super_2 2074907 120 + 9036312 CUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCAC ......(((....)))((.((.(((.((.....(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))......))..))).)).)).. ( -30.80, z-score = -2.38, R) >droWil1.scaffold_180697 1832660 120 + 4168966 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCGAGGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUGGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -35.20, z-score = -2.65, R) >droVir3.scaffold_12823 2414463 120 + 2474545 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUAGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -36.30, z-score = -4.38, R) >droMoj3.scaffold_6496 1760816 120 + 26866924 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUCUCGAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUAGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -34.30, z-score = -3.94, R) >droGri2.scaffold_15245 2652093 120 - 18325388 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCGUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUUGACAUUUUGCACGCGCUAGUUAAGCAC ...........(((((((........)))))))(((((((.(.((((((.((((.((.(((((........))))))).)))))))))).).)))))))........(((.....))).. ( -35.10, z-score = -3.50, R) >anoGam1.chr2L 41834953 120 + 48795086 CUGGAUCCACCGCUUGGCAUCCCGAAACCGAACACGGUCGCGCUGGUGUCGUUCAAUCUGCCCAUUUCCAAGGGGAACAAGAUCCACUGUCUGGACAUACUGCACGCGCUGGUGAAGCAU ..(((((((.....))).))))(((..(((....))))))(((..(((.(((.((.(((.(((.........)))....)))((((.....)))).....)).))))))..)))...... ( -31.30, z-score = 1.66, R) >apiMel3.GroupUn 34969767 120 + 399230636 CUGGAUCCACCAUUAGGGAUUUCGAAGCCCAAUAUGGUUGCACUAGUUAGUUUUAAUUUACCUAUCGCAAAGGGUAAUAAGAUUCAUUGUCUGGACAUUCUGCACGCACUUGUCAAGCAC ........(((((..(((.........)))...)))))(((..((((.(((((((..((((((........))))))))))))).))))....((((...((....))..))))..))). ( -23.60, z-score = 0.59, R) >triCas2.ChLG7 4487171 120 - 17478683 UUAGACCCUCCUUUAGGCAUCUCGAAACCCAAUACCGUUGCUUUAGUGAGCUUCAAUCUGCCCAUCGCCAAGGGCAAUAAAAUCCAUUGUUUAGAUAUUCUACACGCGUUAGUUAAACAC ...((..(((((..(((((...((...........)).))))).)).)))..))....(((((........)))))...........(((((((..((.(.....).))...))))))). ( -20.40, z-score = -0.29, R) >consensus CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCAC ...........((((((..........))))))...(((....((((((.((((.((.(((((........))))))).))))))))))....)))...........(((.....))).. (-22.26 = -21.51 + -0.75)
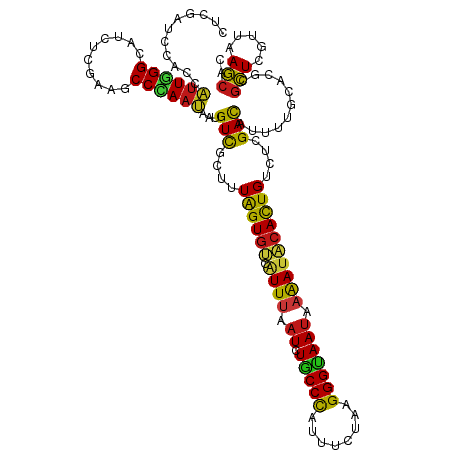
| Location | 20,802,233 – 20,802,353 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Shannon entropy | 0.26500 |
| G+C content | 0.45167 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.21 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20802233 120 - 21146708 GUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))((.((((((((((....(((((((((..((((((........))))))...)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -41.70, z-score = -4.50, R) >droSim1.chr2R 19254849 120 - 19596830 GUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCCUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))((.((((((((((....(((((((((..((((((........))))))...)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -40.50, z-score = -4.03, R) >droSec1.super_23 659237 120 - 989336 GUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCCUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))((.((((((((((....(((((((((..((((((........))))))...)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -40.50, z-score = -4.03, R) >droYak2.chr2R 20765690 120 - 21139217 GUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))((.((((((((((....(((((((((.(((.(((........)))..))).)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -38.30, z-score = -3.25, R) >droEre2.scaffold_4845 22172305 120 - 22589142 GUGCUUAACCAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....)))))((.((((((((((....(((((((((.(((.(((........)))..))).)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -37.70, z-score = -3.02, R) >droAna3.scaffold_13266 12090645 120 - 19884421 GUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCUUCGAAAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))..........((((...(((((((((..((((((........))))))...)))).))))).......(((((((((((........)))))))))))..)))). ( -42.90, z-score = -3.72, R) >dp4.chr3 1892126 120 - 19779522 GUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))..........((((...(((((((((.(((.(((........)))..))).)))).))))).......(((((((.(((........))).)))))))..)))). ( -31.00, z-score = -1.02, R) >droPer1.super_2 2074907 120 - 9036312 GUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))..........((((...(((((((((.(((.(((........)))..))).)))).))))).......(((((((.(((........))).)))))))..)))). ( -31.00, z-score = -1.02, R) >droWil1.scaffold_180697 1832660 120 - 4168966 GUGCUUAACCAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCCUCGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....)))))..........((((...(((((((((..((((((........))))))...)))).))))).......(((((((((((........)))))))))))..)))). ( -41.10, z-score = -3.60, R) >droVir3.scaffold_12823 2414463 120 - 2474545 GUGCUUAACUAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....)))))((.((((((((((....(((((((((..((((((........))))))...)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -43.80, z-score = -4.50, R) >droMoj3.scaffold_6496 1760816 120 - 26866924 GUGCUUAACUAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUCGAGAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....)))))((.((((((((((....(((((((((..((((((........))))))...)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -41.80, z-score = -4.13, R) >droGri2.scaffold_15245 2652093 120 + 18325388 GUGCUUAACUAGCGCGUGCAAAAUGUCAAGACAGUGUAUUUUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAACGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....))))).(((.....((....))(((((((((..((((((........))))))...)))).)))))...))).(((((((((((........)))))))))))....... ( -41.40, z-score = -3.70, R) >anoGam1.chr2L 41834953 120 - 48795086 AUGCUUCACCAGCGCGUGCAGUAUGUCCAGACAGUGGAUCUUGUUCCCCUUGGAAAUGGGCAGAUUGAACGACACCAGCGCGACCGUGUUCGGUUUCGGGAUGCCAAGCGGUGGAUCCAG ....((((((.((((((.((((.((((((..(((.(((......))).).))....)))))).))))..(((.((((((((....))))).))).)))..))))...))))))))..... ( -38.70, z-score = 0.34, R) >apiMel3.GroupUn 34969767 120 - 399230636 GUGCUUGACAAGUGCGUGCAGAAUGUCCAGACAAUGAAUCUUAUUACCCUUUGCGAUAGGUAAAUUAAAACUAACUAGUGCAACCAUAUUGGGCUUCGAAAUCCCUAAUGGUGGAUCCAG ((.((.((((..((....))...)))).))))..((.((((..........((((.(((...............))).))))(((((...(((.........)))..))))))))).)). ( -19.96, z-score = 1.57, R) >triCas2.ChLG7 4487171 120 + 17478683 GUGUUUAACUAACGCGUGUAGAAUAUCUAAACAAUGGAUUUUAUUGCCCUUGGCGAUGGGCAGAUUGAAGCUCACUAAAGCAACGGUAUUGGGUUUCGAGAUGCCUAAAGGAGGGUCUAA (((((.....)))))((.(((.....))).))..((((((((.(((((...)))))((((((..((((((((((.((..(...)..)).))))))))))..))))))....)))))))). ( -36.60, z-score = -2.42, R) >consensus GUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....))))).........(((.....(((((..((((((((((........))))))..))))..)))))..........((((((((........))))))))...))).... (-26.46 = -26.21 + -0.25)
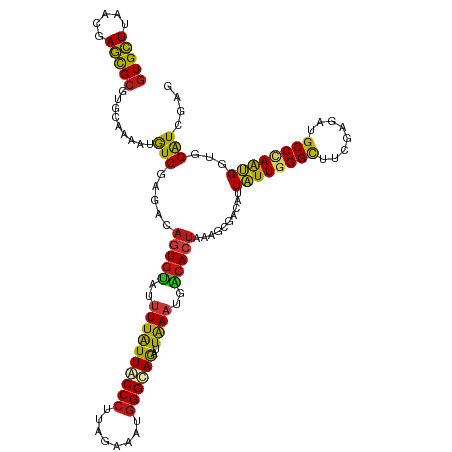
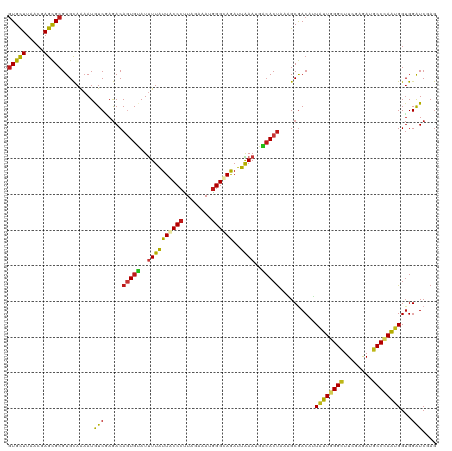
| Location | 20,802,273 – 20,802,389 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Shannon entropy | 0.27925 |
| G+C content | 0.43577 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -26.10 |
| Energy contribution | -25.67 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.48 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
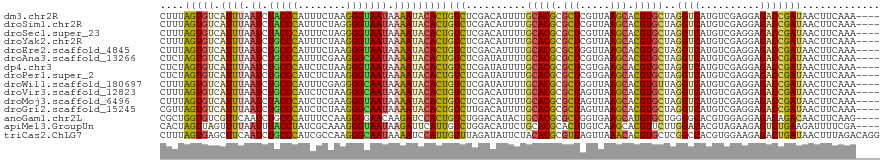
>dm3.chr2R 20802273 116 + 21146708 CUUUAGUGUCAUUUAAUCUACCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -36.20, z-score = -4.31, R) >droSim1.chr2R 19254889 116 + 19596830 CUUUAGUGUCAUUUAAUCUACCCAUUUCUAGGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -36.10, z-score = -3.93, R) >droSec1.super_23 659277 116 + 989336 CUUUAGUGUCAUUUAAUCUACCCAUUUCUAGGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -36.10, z-score = -3.93, R) >droYak2.chr2R 20765730 116 + 21139217 CUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -36.20, z-score = -3.85, R) >droEre2.scaffold_4845 22172345 116 + 22589142 CUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUGGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -37.40, z-score = -3.91, R) >droAna3.scaffold_13266 12090685 116 + 19884421 CUCUAGUGUCAUUUAAUCUGCCCAUUUCGAAGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- (((((((((.((((.((.(((((........))))))).)))))))))).((((((((...(((((.(((.....))).)))))......)))))))))))...............---- ( -37.80, z-score = -3.38, R) >dp4.chr3 1892166 116 + 19779522 CUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- (((((((((.((((.((.(((((........))))))).)))))))))).((((((((...(((((.(((.....))).)))))......)))))))))))...............---- ( -35.80, z-score = -3.24, R) >droPer1.super_2 2074947 116 + 9036312 CUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- (((((((((.((((.((.(((((........))))))).)))))))))).((((((((...(((((.(((.....))).)))))......)))))))))))...............---- ( -35.80, z-score = -3.24, R) >droWil1.scaffold_180697 1832700 116 + 4168966 CUUUAGUGUCAUUUAAUCUGCCCAUUUCGAGGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUGGUUAAGCACGUGUUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -35.80, z-score = -2.88, R) >droVir3.scaffold_12823 2414503 116 + 2474545 CUUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUAGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -38.50, z-score = -4.40, R) >droMoj3.scaffold_6496 1760856 116 + 26866924 CUUUAGUGUCAUUUAAUCUACCCAUCUCGAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUAGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......))))))))).................---- ( -36.50, z-score = -3.94, R) >droGri2.scaffold_15245 2652133 116 - 18325388 CGUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUUGACAUUUUGCACGCGCUAGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA---- .(.((((((.((((.((.(((((........))))))).)))))))))).).((((.....(((((.(((.....))).)))))...))))(((((.(....))))))........---- ( -38.40, z-score = -4.01, R) >anoGam1.chr2L 41834993 116 + 48795086 CGCUGGUGUCGUUCAAUCUGCCCAUUUCCAAGGGGAACAAGAUCCACUGUCUGGACAUACUGCACGCGCUGGUGAAGCAUGUGCUGGGGCACGUGGAGGAGACAGACAACUUCAAG---- .((((.(.((.((((...(((((...((((.(((((......))).))...))))....(.(((((.(((.....))).))))).))))))..)))).)).)))).).........---- ( -37.40, z-score = -0.01, R) >apiMel3.GroupUn 34969807 116 + 399230636 CACUAGUUAGUUUUAAUUUACCUAUCGCAAAGGGUAAUAAGAUUCAUUGUCUGGACAUUCUGCACGCACUUGUCAAGCACGUUCUUGGACACGUAGAAGAGUCUGAAGAUUUUCGA---- .....(..((((((...((((((........))))))..((((((..(((....)))((((((..((.........))..(((....)))..)))))))))))).))))))..)..---- ( -23.30, z-score = 0.38, R) >triCas2.ChLG7 4487211 120 - 17478683 CUUUAGUGAGCUUCAAUCUGCCCAUCGCCAAGGGCAAUAAAAUCCAUUGUUUAGAUAUUCUACACGCGUUAGUUAAACACGUGCUCGGCCACGUGGAAGAGACUGAUAACUUUAGACAGG ((..(((..((.......(((((........)))))............((.(((.....))).))))(((((((...((((((......)))))).....))))))).)))..))..... ( -28.20, z-score = -0.67, R) >consensus CUUUAGUGUCAUUUAAUCUGCCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAA____ ....(((((.((((.((.(((((........))))))).)))))))))(((..........(((((.(((.....))).)))))..((((..........)))))))............. (-26.10 = -25.67 + -0.42)
| Location | 20,802,273 – 20,802,389 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.00 |
| Shannon entropy | 0.27925 |
| G+C content | 0.43577 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -27.07 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.46 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
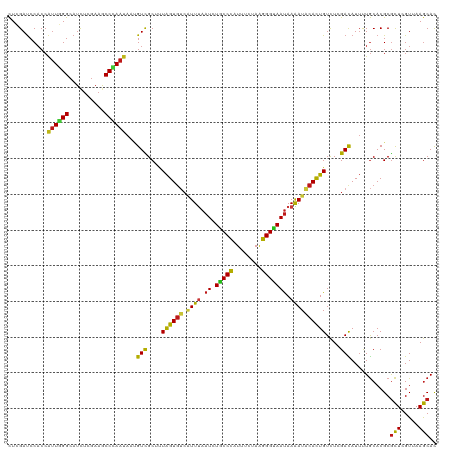
| SVM RNA-class probability | 0.997646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
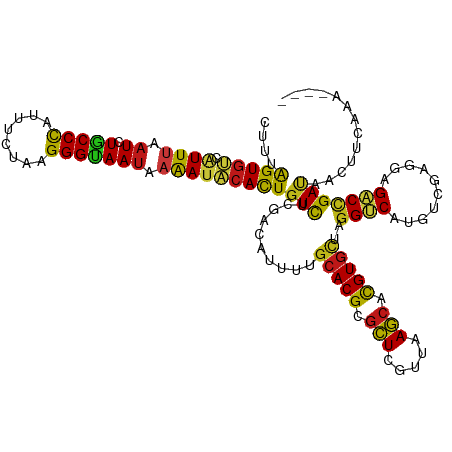
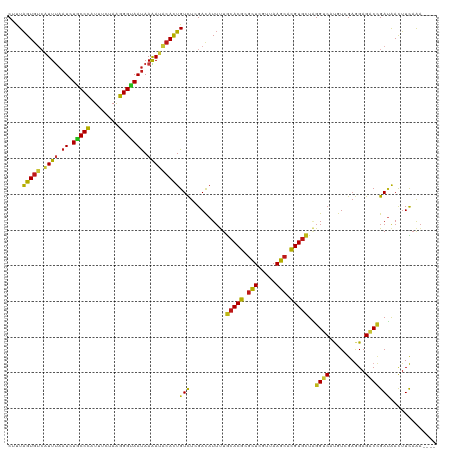
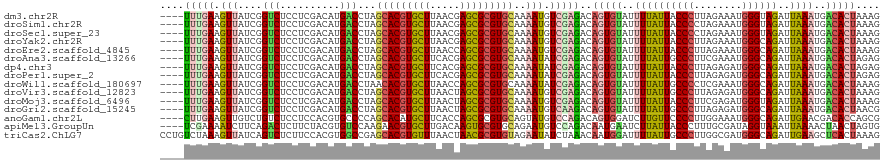
>dm3.chr2R 20802273 116 - 21146708 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGUAGAUUAAAUGACACUAAAG ----..................((((((((......((((((((((...))))))))))...))))))))..(((((((((..((((((........))))))...)))).))))).... ( -40.80, z-score = -5.23, R) >droSim1.chr2R 19254889 116 - 19596830 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCCUAGAAAUGGGUAGAUUAAAUGACACUAAAG ----..................((((((((......((((((((((...))))))))))...))))))))..(((((((((..((((((........))))))...)))).))))).... ( -39.60, z-score = -4.90, R) >droSec1.super_23 659277 116 - 989336 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCCUAGAAAUGGGUAGAUUAAAUGACACUAAAG ----..................((((((((......((((((((((...))))))))))...))))))))..(((((((((..((((((........))))))...)))).))))).... ( -39.60, z-score = -4.90, R) >droYak2.chr2R 20765730 116 - 21139217 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAG ----..................((((((((......((((((((((...))))))))))...))))))))..(((((((((.(((.(((........)))..))).)))).))))).... ( -37.40, z-score = -4.06, R) >droEre2.scaffold_4845 22172345 116 - 22589142 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACCAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAG ----..................((((((((......(((((((((.....)))))))))...))))))))..(((((((((.(((.(((........)))..))).)))).))))).... ( -36.80, z-score = -4.09, R) >droAna3.scaffold_13266 12090685 116 - 19884421 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCUUCGAAAUGGGCAGAUUAAAUGACACUAGAG ----(((((.(((...((((..........))))..((((((((((...))))))))))..))).)))))..(((((((((..((((((........))))))...)))).))))).... ( -39.50, z-score = -3.93, R) >dp4.chr3 1892166 116 - 19779522 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAG ----(((((.(((...((((..........))))..((((((((((...))))))))))..))).)))))..(((((((((.(((.(((........)))..))).)))).))))).... ( -34.10, z-score = -2.63, R) >droPer1.super_2 2074947 116 - 9036312 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAG ----(((((.(((...((((..........))))..((((((((((...))))))))))..))).)))))..(((((((((.(((.(((........)))..))).)))).))))).... ( -34.10, z-score = -2.63, R) >droWil1.scaffold_180697 1832700 116 - 4168966 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAACACGUGCUUAACCAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCCUCGAAAUGGGCAGAUUAAAUGACACUAAAG ----(((((.(((...((((..........))))...((((((((.....))))))))...))).)))))..(((((((((..((((((........))))))...)))).))))).... ( -33.30, z-score = -3.25, R) >droVir3.scaffold_12823 2414503 116 - 2474545 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACUAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAAAG ----..................((((((((......(((((((((.....)))))))))...))))))))..(((((((((..((((((........))))))...)))).))))).... ( -42.90, z-score = -5.24, R) >droMoj3.scaffold_6496 1760856 116 - 26866924 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACUAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUCGAGAUGGGUAGAUUAAAUGACACUAAAG ----..................((((((((......(((((((((.....)))))))))...))))))))..(((((((((..((((((........))))))...)))).))))).... ( -40.90, z-score = -4.92, R) >droGri2.scaffold_15245 2652133 116 + 18325388 ----UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACUAGCGCGUGCAAAAUGUCAAGACAGUGUAUUUUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAACG ----(((((.(((...((((..........))))..(((((((((.....)))))))))..))).)))))..(((((((((..((((((........))))))...)))).))))).... ( -39.90, z-score = -4.42, R) >anoGam1.chr2L 41834993 116 - 48795086 ----CUUGAAGUUGUCUGUCUCCUCCACGUGCCCCAGCACAUGCUUCACCAGCGCGUGCAGUAUGUCCAGACAGUGGAUCUUGUUCCCCUUGGAAAUGGGCAGAUUGAACGACACCAGCG ----......((((..((((.....(((((((...(((....)))......)))))))((((.((((((..(((.(((......))).).))....)))))).))))...)))).)))). ( -34.80, z-score = -0.58, R) >apiMel3.GroupUn 34969807 116 - 399230636 ----UCGAAAAUCUUCAGACUCUUCUACGUGUCCAAGAACGUGCUUGACAAGUGCGUGCAGAAUGUCCAGACAAUGAAUCUUAUUACCCUUUGCGAUAGGUAAAUUAAAACUAACUAGUG ----..(((....))).........(((.((((.....(((..((.....))..)))(((((..((..(((.......)))....))..))))))))).))).................. ( -16.90, z-score = 1.29, R) >triCas2.ChLG7 4487211 120 + 17478683 CCUGUCUAAAGUUAUCAGUCUCUUCCACGUGGCCGAGCACGUGUUUAACUAACGCGUGUAGAAUAUCUAAACAAUGGAUUUUAUUGCCCUUGGCGAUGGGCAGAUUGAAGCUCACUAAAG .........((((.(((((((...((.(((.((((((((((((((.....))))))))...((.(((((.....))))).))......)))))).))))).)))))))))))........ ( -35.00, z-score = -2.33, R) >consensus ____UUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAG ....(((((.(((....(((..........)))...(((((((((.....)))))))))..))).)))))..(((((..((((((((((........))))))..))))..))))).... (-27.07 = -27.30 + 0.23)
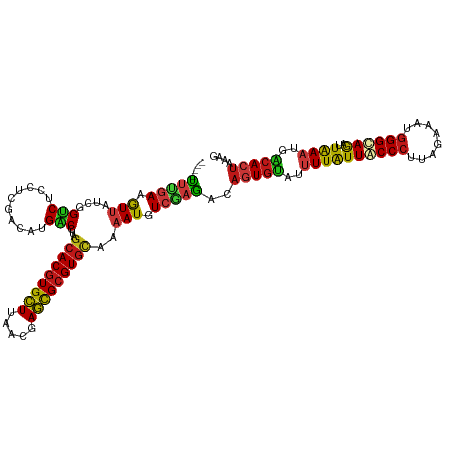
| Location | 20,802,389 – 20,802,505 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Shannon entropy | 0.23459 |
| G+C content | 0.46450 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -28.39 |
| Energy contribution | -28.29 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
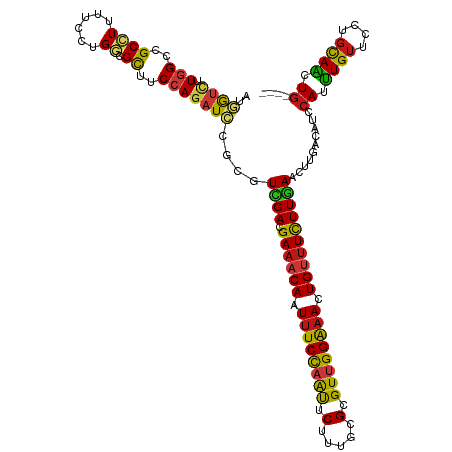
>dm3.chr2R 20802389 116 + 21146708 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -37.80, z-score = -2.76, R) >droSim1.chr2R 19255005 116 + 19596830 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -37.80, z-score = -2.76, R) >droSec1.super_23 659393 116 + 989336 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -37.80, z-score = -2.76, R) >droYak2.chr2R 20765846 116 + 21139217 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -37.80, z-score = -2.76, R) >droEre2.scaffold_4845 22172461 116 + 22589142 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAACGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -38.90, z-score = -3.31, R) >droAna3.scaffold_13266 12090801 116 + 19884421 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUUAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----.....((..((.((......))))..(((...((((((((((((((..(.....)..))))))))))))))...)))))((.(((((...((((.......)))).)).))))).. ( -35.80, z-score = -2.37, R) >dp4.chr3 1892282 116 + 19779522 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -37.80, z-score = -2.76, R) >droPer1.super_2 2075063 116 + 9036312 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -37.80, z-score = -2.76, R) >droWil1.scaffold_180697 1832816 116 + 4168966 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGUAAAGAAUUGGAAAUUGUUUCAUCGACAAGGAUCUGGAAGCGUCAGGAGAAGGCGGCCAAAACGAU ----..((((..........(((((.....))))).((((((((((((((..(.....)..))))))))))))))..)))).......(((..((.((....))..))..)))....... ( -30.20, z-score = -1.92, R) >droVir3.scaffold_12823 2414619 116 + 2474545 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACACGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCAGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((....(((.......)))..)).))))).. ( -33.70, z-score = -2.30, R) >droMoj3.scaffold_6496 1760972 116 + 26866924 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACACGAAAAGAAUUGGAAAUUGUUUCAUCAACACGGAUCUGGAAGCGCCAGGAAAAGGCAGCAAAGACCAU ----..(((((..((.....(((((.....))))).((((((((((((((..(.....)..)))))))))))))).))........(((((.....))))).....)))))......... ( -32.70, z-score = -3.40, R) >droGri2.scaffold_15245 2652249 116 - 18325388 ----CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ----..(((((.........(((((.....))))).((((((((((((((..(.....)..))))))))))))))).))))..((.(((((...((((.......)))).)).))))).. ( -37.80, z-score = -2.76, R) >anoGam1.chr2L 41835109 116 + 48795086 ----CAGCUACAAGACCAGAUGGACCAGAAGUUCAAAAAGCAGUUCCCAACGCGAAAGGAGCUGGAGAUUGUGUCGUCGACGCGCAUCUGGAAGCGACAGGAGAAGGCGGCCAAAACGAU ----..(((......(((((((..................(((((((..........)))))))......(((((...))))).)))))))..((...........)))))......... ( -27.80, z-score = 0.55, R) >apiMel3.GroupUn 34969923 116 + 399230636 ----AAGCUGCAGGAACAAAUGGAUAUUAAGUUUAAAAAACAAUUUCCCACUCGUAAAGAAUUAGAAAUUGUUUCUUCAACGAGAAUGUGGAAACGUCAAGAUAAAGCAGCUAGGUUGAU ----.((((((............(((((..(((.((.((((((((((....((.....))....)))))))))).)).)))...)))))(....)...........))))))........ ( -25.50, z-score = -2.06, R) >triCas2.ChLG7 4487371 120 - 17478683 CUUCCAGUUACAAGAACAAAUGGAUAUCAAAUUCAAGAAGCAGUUUCCGACGCGAAAAGAGCUGGAAAUUGUUUCCUCGACUCGGAUUUGGAAACGGCAAGAUAAGGCCGCCAGGUUGAU ..(((((((.....)))...))))............(((((((((((((..((.......))))))))))))))).(((((..((....(....)(((........))).))..))))). ( -33.90, z-score = -1.88, R) >consensus ____CAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAU ......(((((..............(((..(((...((((((((((((((..(.....)..))))))))))))))...)))...)))((((.....))))......)))))......... (-28.39 = -28.29 + -0.10)
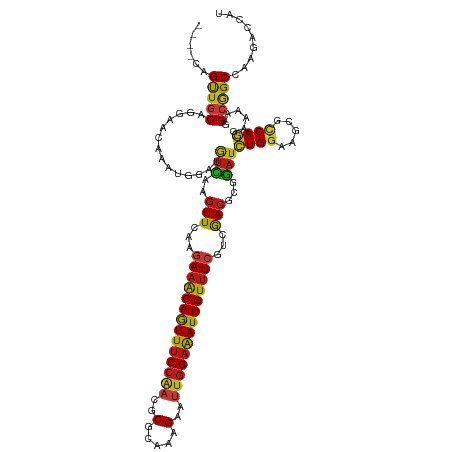
| Location | 20,802,389 – 20,802,505 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Shannon entropy | 0.23459 |
| G+C content | 0.46450 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -22.19 |
| Energy contribution | -22.03 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20802389 116 - 21146708 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.93, R) >droSim1.chr2R 19255005 116 - 19596830 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.93, R) >droSec1.super_23 659393 116 - 989336 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.93, R) >droYak2.chr2R 20765846 116 - 21139217 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.93, R) >droEre2.scaffold_4845 22172461 116 - 22589142 AUGGUCUUGGCCGCCUUUUCCUGGCGUUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.91, R) >droAna3.scaffold_13266 12090801 116 - 19884421 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUAAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))..(((((.((((((.((((((((.(.....).)))))))).))))))......)))))...((.((((....)))).))---- ( -31.20, z-score = -2.61, R) >dp4.chr3 1892282 116 - 19779522 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.93, R) >droPer1.super_2 2075063 116 - 9036312 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.93, R) >droWil1.scaffold_180697 1832816 116 - 4168966 AUCGUUUUGGCCGCCUUCUCCUGACGCUUCCAGAUCCUUGUCGAUGAAACAAUUUCCAAUUCUUUACGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- .....(((((..((..((....)).))..)))))....((((((.((((((.((((((((.(.....).)))))))).))))))......))))))..((.((((....)))).))---- ( -25.40, z-score = -2.52, R) >droVir3.scaffold_12823 2414619 116 - 2474545 AUGGUCUUGGCUGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGUGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..(((((.(((.(((.......))))))...))))).((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -32.60, z-score = -3.00, R) >droMoj3.scaffold_6496 1760972 116 - 26866924 AUGGUCUUUGCUGCCUUUUCCUGGCGCUUCCAGAUCCGUGUUGAUGAAACAAUUUCCAAUUCUUUUCGUGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- (((((((..((.(((.......)))))....))).))))((((..((((((.((((((((.(.....).)))))))).))))))..((((.((.....))...))))...))))..---- ( -31.00, z-score = -3.76, R) >droGri2.scaffold_15245 2652249 116 + 18325388 AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG---- ..((..((((.((((.......))))...))))..))((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)).....---- ( -33.40, z-score = -2.93, R) >anoGam1.chr2L 41835109 116 - 48795086 AUCGUUUUGGCCGCCUUCUCCUGUCGCUUCCAGAUGCGCGUCGACGACACAAUCUCCAGCUCCUUUCGCGUUGGGAACUGCUUUUUGAACUUCUGGUCCAUCUGGUCUUGUAGCUG---- ......................((..(..(((((((.(((((...))).(((....(((.((((........)))).)))....)))........)).)))))))....)..))..---- ( -24.50, z-score = 1.24, R) >apiMel3.GroupUn 34969923 116 - 399230636 AUCAACCUAGCUGCUUUAUCUUGACGUUUCCACAUUCUCGUUGAAGAAACAAUUUCUAAUUCUUUACGAGUGGGAAAUUGUUUUUUAAACUUAAUAUCCAUUUGUUCCUGCAGCUU---- ........((((((........((((............)))).((((((((((((((.((((.....)))).)))))))))))))).......................)))))).---- ( -26.90, z-score = -3.72, R) >triCas2.ChLG7 4487371 120 + 17478683 AUCAACCUGGCGGCCUUAUCUUGCCGUUUCCAAAUCCGAGUCGAGGAAACAAUUUCCAGCUCUUUUCGCGUCGGAAACUGCUUCUUGAAUUUGAUAUCCAUUUGUUCUUGUAACUGGAAG .((((...((((((........)))((((((..((.((((..((((((.....))))...))..)))).)).)))))).)))..))))........((((.(((......))).)))).. ( -25.90, z-score = -0.11, R) >consensus AUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUG____ ..((((.(((..((((......)).))..))))))).((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...))......... (-22.19 = -22.03 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:27 2011