
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,902,659 – 3,902,771 |
| Length | 112 |
| Max. P | 0.894676 |

| Location | 3,902,659 – 3,902,771 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.50 |
| Shannon entropy | 0.43536 |
| G+C content | 0.44734 |
| Mean single sequence MFE | -36.21 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3902659 112 + 23011544 GGCCCAAUAUUUUGGGCUUAAUUGUGGCACGAAUACCCCAUUGCAGAAGGGCUAACAAUGUGA---UGGCUAUGACCAUCAUAGCC---GGAUAGAGCUCUUCCAUUGGAUAC-ACAAU (((((((....)))))))..((((((....(.....)(((.((..((((((((.....(((..---((((((((.....)))))))---).))).)))))))))).)))...)-))))) ( -40.10, z-score = -3.38, R) >droSim1.chr2L 3857799 112 + 22036055 GGCCCGCUAUUUUGGGCUUAAUUGUAGCACGAAUACCCCAUUGCAGAAGGGCUAACAAUGUGA---UGGCUAUGACCAUCAUAGCC---GGAUUGAGCUCUUCCAUUGGGUAC-AUAAU ((((((......))))))..................((((.((..((((((((..((((....---((((((((.....)))))))---).)))))))))))))).))))...-..... ( -38.50, z-score = -2.46, R) >droSec1.super_5 2005619 112 + 5866729 GGCCCGCUAUUUUGGGCUUAAUUGUAGCACGAAUACCCCAUUGCAGAAGGGCUAACAAUGUGA---UGGCUAUGACCAUCAUAGCC---GGAUUGAGCUCUUCCAUUGGGUAC-AUAAU ((((((......))))))..................((((.((..((((((((..((((....---((((((((.....)))))))---).)))))))))))))).))))...-..... ( -38.50, z-score = -2.46, R) >droYak2.chr2L 3910989 112 + 22324452 GGCCCGAUAUUUUGGGCUUAAUUGCAACACGAAUGCCCCAAUGCAGAAGGGCUAACAAUGUGA---UAGCAAUGACCAUAAGAGCU---GGAUAGAGCUCUUCCAUUGCGUAA-AUGGU (((((((....)))))))...((((..((((...((((..........))))......)))).---..((((((.....(((((((---......))))))).))))))))))-..... ( -35.30, z-score = -2.13, R) >droEre2.scaffold_4929 3954895 112 + 26641161 GGCCCGAUAUUUUGGGCUUAAUUGCAACACGAAUAGCCCAUUGCAGAAGGGCCAACAAUGUGA---UAGCCAUGACCAUAAGAGUU---GGAUAGAGCUCUUCCAUUGGGUAC-AUAAU (((((.......((((((...(((.....)))..))))))........)))))..(((((..(---.(((..((.(((.......)---)).))..))).)..))))).....-..... ( -34.56, z-score = -2.22, R) >droAna3.scaffold_12943 443417 118 - 5039921 AACUGGGUAUUGGCAGUAUAGUUGCAACACGAUCGCUGUAGUGCAAUAUCGCCAGAAAUAUGAGAACAGUUAUGAAAAUGGAGACCACUGGAUAGUAUUCAACUAUCCUGUCUGAUAA- ((((((((((((.((.((((((..(.....)...)))))).)))))))))..((......))....)))))..........((((....(((((((.....))))))).)))).....- ( -30.30, z-score = -1.29, R) >consensus GGCCCGAUAUUUUGGGCUUAAUUGCAACACGAAUACCCCAUUGCAGAAGGGCUAACAAUGUGA___UAGCUAUGACCAUCAUAGCC___GGAUAGAGCUCUUCCAUUGGGUAC_AUAAU ((((((......))))))..................((((.((..((((((((.....(((......((((...........)))).....))).)))))))))).))))......... (-16.59 = -16.80 + 0.21)

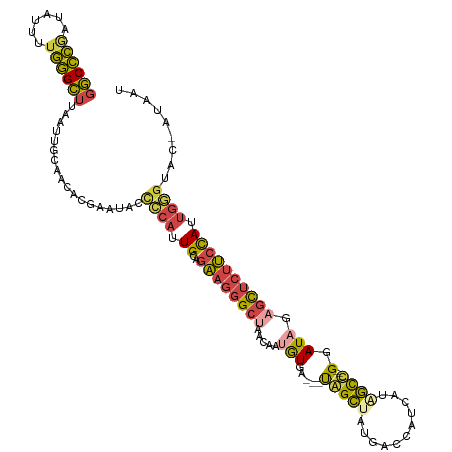
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:53 2011