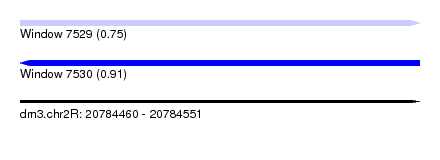
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,784,460 – 20,784,551 |
| Length | 91 |
| Max. P | 0.911012 |

| Location | 20,784,460 – 20,784,551 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 61.39 |
| Shannon entropy | 0.68423 |
| G+C content | 0.66221 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -14.83 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
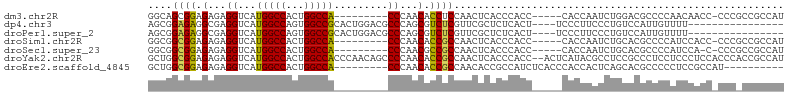
>dm3.chr2R 20784460 91 + 21146708 AUGGCGGCGGG-GGUUGUUGGGGCGUCCAGAUUGGUG-----GGUGGGUGAGUUGGAGGUGUUGGG---------UGGCCAGUGGCCAUGACCUCUCUCCGCUGCC ..(((((((((-((...(..(.((.(((((.(((.(.-----....).))).))))).)).)..)(---------(((((...))))))......))))))))))) ( -38.50, z-score = -1.20, R) >dp4.chr3 1872285 86 + 19779522 ----------------AAAACAAUGGACAGGGAAGGGA----AGUGAGAGCGAACGAGACGCUGGGCGUCCAGUGCGGCCACUGGCCAUGACCUCGCCUCUCCGCU ----------------.............((..(((..----.((....))...((((.((((((....)))))).((((...)))).....)))))))..))... ( -29.50, z-score = -0.17, R) >droPer1.super_2 2055329 86 + 9036312 ----------------AAAACAAUGGACAGGGAAGGGA----AGUGAGAGCGAACGAGACGCUGGGCGUCCAGUGCGGCCACUGGCCAUGACCUCGCCUCUCCGCU ----------------.............((..(((..----.((....))...((((.((((((....)))))).((((...)))).....)))))))..))... ( -29.50, z-score = -0.17, R) >droSim1.chr2R 19236985 91 + 19596830 AUGGCGGCGGG-GGUGGAUGGGGCGUGCAGAUUGGUG-----GGUGGGUGAGUUGGCGGUGUUGGG---------UGGCCAGUGGCCAUGACCUCUCUCCGCCGCC ..(((((((((-((.((((((.((.(((......)))-----.)).(((.(.(..((...))..).---------).))).....))))..)).).)))))))))) ( -38.60, z-score = -0.46, R) >droSec1.super_23 641475 90 + 989336 AUGGCGGCGGG-G-UGGAUGGGGCGUGCAGAUUGGUG-----GGUGGGUGAGUUGGCGGCGUUGGG---------UGGCCAGUGGCCAUGACCUCUCUCCGCCGCC ..(((((((((-(-.(((.((.((.(((......)))-----.)).(((.(.(((((.((.....)---------).)))))).)))....))))))))))))))) ( -42.80, z-score = -1.67, R) >droYak2.chr2R 20747447 104 + 21139217 AUGGCGGUGGGUGGAGGGAGGAGGGCGGAGGCGUAUGAGU--GGUGGGUGAGUUGGCGGUGUUGGGGCUGUUGGGUGGCCAGUGGCCAUGACCUCUCUCCGCCAGC ...((....((((((((((((...(((....)))....((--(((.(((.(.(..(((((......)))))..).).)))....)))))..)))))))))))).)) ( -46.80, z-score = -3.57, R) >droEre2.scaffold_4845 22154267 87 + 22589142 ----------AUGGCGGAGGGGGCGUGCUGAGUGGUGGGUGAGAUGGCGGUGUUGGCGGUGUUGGG---------UGGCCAGUGGCCAUGACCUCUCUCCGCCAGC ----------.((((((((((((((((.(.(.((((.(.(.((((.((.......)).)).)).).---------).)))).).).)))).)).)))))))))).. ( -38.90, z-score = -1.59, R) >consensus AUGGCGG_GGG_GGUGGAAGGGGCGUGCAGAGUGGUG_____GGUGGGUGAGUUGGCGGUGUUGGG_________UGGCCAGUGGCCAUGACCUCUCUCCGCCGCC ......................................................(((((.(..(((.........(((((...)))))...)))..).)))))... (-14.83 = -14.73 + -0.10)


| Location | 20,784,460 – 20,784,551 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 61.39 |
| Shannon entropy | 0.68423 |
| G+C content | 0.66221 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -11.09 |
| Energy contribution | -10.30 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20784460 91 - 21146708 GGCAGCGGAGAGAGGUCAUGGCCACUGGCCA---------CCCAACACCUCCAACUCACCCACC-----CACCAAUCUGGACGCCCCAACAACC-CCCGCCGCCAU (((.((((.(.(((((..(((((...)))))---------......))))))............-----........(((.....)))......-.)))).))).. ( -27.70, z-score = -2.51, R) >dp4.chr3 1872285 86 - 19779522 AGCGGAGAGGCGAGGUCAUGGCCAGUGGCCGCACUGGACGCCCAGCGUCUCGUUCGCUCUCACU----UCCCUUCCCUGUCCAUUGUUUU---------------- ...(((.(((.((((....((((...))))((((.((((((...)))))).))..)).......----..)))).))).)))........---------------- ( -30.90, z-score = -0.85, R) >droPer1.super_2 2055329 86 - 9036312 AGCGGAGAGGCGAGGUCAUGGCCAGUGGCCGCACUGGACGCCCAGCGUCUCGUUCGCUCUCACU----UCCCUUCCCUGUCCAUUGUUUU---------------- ...(((.(((.((((....((((...))))((((.((((((...)))))).))..)).......----..)))).))).)))........---------------- ( -30.90, z-score = -0.85, R) >droSim1.chr2R 19236985 91 - 19596830 GGCGGCGGAGAGAGGUCAUGGCCACUGGCCA---------CCCAACACCGCCAACUCACCCACC-----CACCAAUCUGCACGCCCCAUCCACC-CCCGCCGCCAU ((((((((.(((.(((....)))..((((..---------.........)))).))).......-----((......))...............-.)))))))).. ( -28.30, z-score = -1.97, R) >droSec1.super_23 641475 90 - 989336 GGCGGCGGAGAGAGGUCAUGGCCACUGGCCA---------CCCAACGCCGCCAACUCACCCACC-----CACCAAUCUGCACGCCCCAUCCA-C-CCCGCCGCCAU ((((((((.(.(.((....(((....(((..---------......)))((.............-----.........))..)))....)).-)-))))))))).. ( -29.25, z-score = -1.86, R) >droYak2.chr2R 20747447 104 - 21139217 GCUGGCGGAGAGAGGUCAUGGCCACUGGCCACCCAACAGCCCCAACACCGCCAACUCACCCACC--ACUCAUACGCCUCCGCCCUCCUCCCUCCACCCACCGCCAU ..((((((.(((.(((..(((((...))))).......((.........))..........)))--.)))....((....)).................)))))). ( -23.90, z-score = -1.18, R) >droEre2.scaffold_4845 22154267 87 - 22589142 GCUGGCGGAGAGAGGUCAUGGCCACUGGCCA---------CCCAACACCGCCAACACCGCCAUCUCACCCACCACUCAGCACGCCCCCUCCGCCAU---------- ..((((((((.(.(((..(((((...)))))---------.........((.......))......................))).))))))))).---------- ( -25.70, z-score = -1.73, R) >consensus GGCGGCGGAGAGAGGUCAUGGCCACUGGCCA_________CCCAACACCGCCAACUCACCCACC_____CACCAAUCUGCACGCCCCAUCCACC_CCC_CCGCCAU ....((((.(...((...(((((...))))).........))...).))))....................................................... (-11.09 = -10.30 + -0.79)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:18 2011