
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,746,717 – 20,746,864 |
| Length | 147 |
| Max. P | 0.866122 |

| Location | 20,746,717 – 20,746,826 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Shannon entropy | 0.30319 |
| G+C content | 0.37228 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
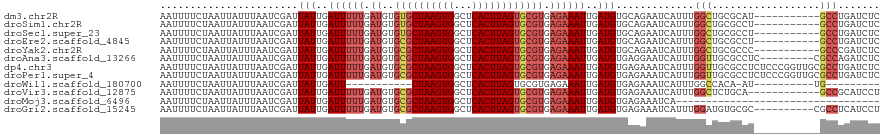
>dm3.chr2R 20746717 109 - 21146708 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGUGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCAGAAUCAUUUGGCUGCGCAU-----------GCCUGAUCUC ....................((((((..((((((.((((..((((((((...)))))))))))).))))))..)(((((((..........))))))).-----------...))))).. ( -29.20, z-score = -1.98, R) >droSim1.chr2R 19198260 109 - 19596830 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGUGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCAGAAUCAUUUGGCUGCGCCU-----------GCCUGAUCUC ....................((((((..((((((.((((..((((((((...)))))))))))).))))))..))((((((..........))))))..-----------....)))).. ( -27.80, z-score = -1.85, R) >droSec1.super_23 603182 109 - 989336 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGUGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCAGAAUCAUUUGGCUGCGCCU-----------GCCUGAUCUC ....................((((((..((((((.((((..((((((((...)))))))))))).))))))..))((((((..........))))))..-----------....)))).. ( -27.80, z-score = -1.85, R) >droEre2.scaffold_4845 22111594 109 - 22589142 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCAGAAUCAUUUGGCUGCGCCU-----------GCCUGAUCUC ....................((((((..((((((.((..((((((((((...)))))))))))).))))))..))((((((..........))))))..-----------....)))).. ( -29.20, z-score = -1.98, R) >droYak2.chr2R 20709063 109 - 21139217 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCAGAAUCAUUUGGCUGCGCCC-----------GCCCGAUCUC .................((((...((..((((((.((..((((((((((...)))))))))))).))))))..))((((((..........))))))..-----------...))))... ( -29.70, z-score = -2.00, R) >droAna3.scaffold_13266 12033430 111 - 19884421 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGGAAUCAUUUGGUUGCGCCUC---------CGCCAGAUCUC ....................((((((..((((((.((..((((((((((...)))))))))))).))))))..))(.((((...((......))..))))---------)....)))).. ( -27.80, z-score = -1.28, R) >dp4.chr3 9016029 120 + 19779522 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGUUGCGCCUCUCCCGGUUGCGCCUGAUCUC ....................((((((..((((((.((..((((((((((...)))))))))))).))))))..))((((....))))..(((.(((((......)).)))))).)))).. ( -32.30, z-score = -2.22, R) >droPer1.super_4 1075937 120 + 7162766 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGUUGCGCCUCUCCCGGUUGCGCCUGAUCUC ....................((((((..((((((.((..((((((((((...)))))))))))).))))))..))((((....))))..(((.(((((......)).)))))).)))).. ( -32.30, z-score = -2.22, R) >droWil1.scaffold_180700 249064 89 - 6630534 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUU-----------CUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGCCACA-AU----------UG--------- ........................((((...-----------((((((((((((((((((.(....)..))))).)))))...))))))))...))-))----------..--------- ( -16.30, z-score = -0.51, R) >droVir3.scaffold_12875 1500103 108 - 20611582 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGCUCUGCA------------GCCGCAUCCU ...(((((.((((...((((((...))))))..))))..((((((((((...))))))))))...)))))..(((((((....))....((((....)------------)))))))).. ( -30.50, z-score = -2.43, R) >droMoj3.scaffold_6496 4606629 86 - 26866924 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCA---------------------------------- ..................((...(((..((((((.((..((((((((((...)))))))))))).))))))..)))...)).....---------------------------------- ( -22.70, z-score = -2.84, R) >droGri2.scaffold_15245 8825272 110 + 18325388 AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGAUGUGCGC----------CGCCUCAUCCU .......................(((..((((((.((..((((((((((...)))))))))))).))))))..)))(((....)))...(((((.((..----------.))..))))). ( -29.80, z-score = -2.40, R) >consensus AAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGCUGCGCCU___________GCCUGAUCUC .......................(((..((((((.((..((((((((((...)))))))))))).))))))..)))............................................ (-18.58 = -19.08 + 0.50)


| Location | 20,746,746 – 20,746,864 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Shannon entropy | 0.09904 |
| G+C content | 0.30293 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
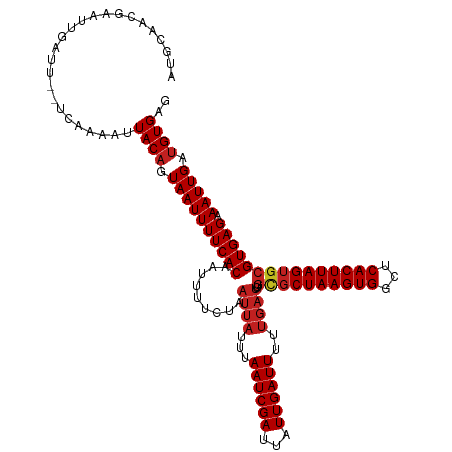
>dm3.chr2R 20746746 118 - 21146708 AUGCAUCGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGUGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCA ..(((((((.(((...--.)))...........(((((((.........((((...((((((...))))))..))))..(..(((((((...)))))))..)))))))).)))))))... ( -28.90, z-score = -2.03, R) >droSim1.chr2R 19198289 118 - 19596830 AUGCAUCGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGUGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCA ..(((((((.(((...--.)))...........(((((((.........((((...((((((...))))))..))))..(..(((((((...)))))))..)))))))).)))))))... ( -28.90, z-score = -2.03, R) >droSec1.super_23 603211 118 - 989336 AUGCAUCGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGUGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCA ..(((((((.(((...--.)))...........(((((((.........((((...((((((...))))))..))))..(..(((((((...)))))))..)))))))).)))))))... ( -28.90, z-score = -2.03, R) >droEre2.scaffold_4845 22111623 118 - 22589142 AUGCAUCGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCA ..(((((((.(((...--.)))...........(((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))))).)))))))... ( -31.30, z-score = -2.74, R) >droYak2.chr2R 20709092 118 - 21139217 AUGCAUCGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCA ..(((((((.(((...--.)))...........(((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))))).)))))))... ( -31.30, z-score = -2.74, R) >droAna3.scaffold_13266 12033461 118 - 19884421 AUGCAUCGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG ..(((((((.(((...--.)))...........(((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))))).)))))))... ( -31.30, z-score = -2.90, R) >dp4.chr3 9016069 118 + 19779522 AUGCAACGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG .......(((.....)--))....(((((.((((((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))).))))).))))). ( -28.90, z-score = -2.23, R) >droPer1.super_4 1075977 118 + 7162766 AUGCAACGAAUUGAUU--UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG .......(((.....)--))....(((((.((((((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))).))))).))))). ( -28.90, z-score = -2.23, R) >droWil1.scaffold_180700 249084 107 - 6630534 AUGCAACGAAUUGAUUU-C-AAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUU-----------CUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG .......(((.....))-)-....(((((.((((((((((................((((((...))))))-----------(((((((...)))))))...))))).))))).))))). ( -18.80, z-score = -0.26, R) >droVir3.scaffold_12875 1500131 119 - 20611582 AUGCAACGAAUUGAUUU-CGAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG ......((((.....))-))....(((((.((((((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))).))))).))))). ( -30.90, z-score = -2.75, R) >droMoj3.scaffold_6496 4606635 119 - 26866924 AUGCAACGAAUUGAUUU-CGAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG ......((((.....))-))....(((((.((((((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))).))))).))))). ( -30.90, z-score = -2.75, R) >droGri2.scaffold_15245 8825302 120 + 18325388 AUGCAACGAAUUGAUUUCUCAAAAUUACUGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG ......((.((..((((((((............................((((...((((((...))))))..))))..((((((((((...)))))))))).))))))))..)).)).. ( -29.60, z-score = -2.54, R) >consensus AUGCAACGAAUUGAUU__UCAAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAG .........................((((.((((((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))).))))).)))).. (-24.02 = -24.52 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:17 2011