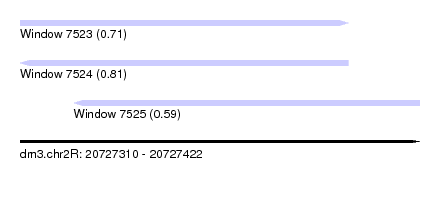
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,727,310 – 20,727,422 |
| Length | 112 |
| Max. P | 0.808980 |

| Location | 20,727,310 – 20,727,402 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.19 |
| Shannon entropy | 0.28170 |
| G+C content | 0.45963 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -17.93 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20727310 92 + 21146708 -----AAAGGGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGUGGACAACUGCCGAGCGGCG--CUGAAACAAUUCGAUUACCACAAG -----...((.......(((((((....)))))))..........((((.((((...((..((((....))))--.))..)))).))))...))..... ( -23.60, z-score = -1.40, R) >droSim1.chr2R 19175384 92 + 19596830 -----AAAGGGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGUGGACAACUGCCGAGCGGCG--CUGAAACAAUUCGAUUACCACAAG -----...((.......(((((((....)))))))..........((((.((((...((..((((....))))--.))..)))).))))...))..... ( -23.60, z-score = -1.40, R) >droSec1.super_23 583805 92 + 989336 -----AAAGGGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGUGGACAACUGCCGAGCGGCG--CUGAAACAAUUCGAUUACCACAAG -----...((.......(((((((....)))))))..........((((.((((...((..((((....))))--.))..)))).))))...))..... ( -23.60, z-score = -1.40, R) >droYak2.chr2R 20688381 92 + 21139217 -----AAAGGGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGUGGACAACUGCCGAGCGGCG--CUGAAACAAUUCGAUUACCACAAG -----...((.......(((((((....)))))))..........((((.((((...((..((((....))))--.))..)))).))))...))..... ( -23.60, z-score = -1.40, R) >droEre2.scaffold_4845 22092002 92 + 22589142 -----AAAGGGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGGUUUGUGGACAACUGCCGAGCGGCG--CUGAAACAAUUCGAUUACCACAAG -----........(((((((((((....))))))).....(((.((((((..((.....)).))))))))).)--)))..................... ( -25.40, z-score = -1.46, R) >droAna3.scaffold_13266 12013407 97 + 19884421 AAAGGGAACGGAGCGGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGCGGACAACUGCCGAGCGGCG--CUGAAACAAUUCGAUUACCACGAG ...((...((((.(((((((((((....))))))).....(((.((((....((((....))))))))))).)--)))......))))....))..... ( -25.70, z-score = -0.64, R) >dp4.chr3 8993621 93 - 19779522 ----UAAAGAGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGCGGACAACUGCCGAGCGGUG--CUGAAACAAUUCGAUUACCACAAA ----.........(((((((((((....)))))))....((((.((((....((((....)))))))))))))--)))..................... ( -23.70, z-score = -2.03, R) >droPer1.super_4 1053578 93 - 7162766 ----UAAAGAGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGCGGACAACUGCCGAGCGGCG--CUGAAACAAUUCGAUUACCACAAA ----.........(((((((((((....))))))).....(((.((((....((((....))))))))))).)--)))..................... ( -22.40, z-score = -1.59, R) >droVir3.scaffold_12875 1473847 87 + 20611582 -----AACAAUAACGACUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGCGGACGGCCGUCGAGCGGCGCUCUGAAACAAUUCGAUUA------- -----..((((......(((((((....)))))))....))))..((((.(((((((..((((.....))))..))))...))).))))...------- ( -24.90, z-score = -1.92, R) >droMoj3.scaffold_6496 4578774 74 + 26866924 -----AACAUUAAUGACUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGCGGACGGCG-------------CUGAAACAAUUCGAUUA------- -----....(((((...(((((((....)))))))....))))).((((.((((((......-------------)))...))).))))...------- ( -15.60, z-score = -1.47, R) >droGri2.scaffold_15245 8803343 85 - 18325388 -----AACAAUAAUAACUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGCGGACGACUGUCGAGCGGCG--CUGAAACAAUUCGAUUA------- -----..((((..(((.(((((((....)))))))))).))))..((((.((((((.((.(((.....)))))--)))...))).))))...------- ( -21.10, z-score = -1.94, R) >consensus _____AAAGGGAACAGCUUGACCUCUGGAGGUCAAUUAAAUUGACUCGAUUUGCGGACAACUGCCGAGCGGCG__CUGAAACAAUUCGAUUACCACAAG .................(((((((....)))))))..........((((.(((........((((....))))........))).)))).......... (-17.93 = -17.73 + -0.20)


| Location | 20,727,310 – 20,727,402 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.19 |
| Shannon entropy | 0.28170 |
| G+C content | 0.45963 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -15.01 |
| Energy contribution | -14.95 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20727310 92 - 21146708 CUUGUGGUAAUCGAAUUGUUUCAG--CGCCGCUCGGCAGUUGUCCACAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCCCUUU----- ...(..((..((((.((((..(((--((((....))).))))...)))).))))..........(((((((....)))))))))..).......----- ( -26.40, z-score = -2.62, R) >droSim1.chr2R 19175384 92 - 19596830 CUUGUGGUAAUCGAAUUGUUUCAG--CGCCGCUCGGCAGUUGUCCACAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCCCUUU----- ...(..((..((((.((((..(((--((((....))).))))...)))).))))..........(((((((....)))))))))..).......----- ( -26.40, z-score = -2.62, R) >droSec1.super_23 583805 92 - 989336 CUUGUGGUAAUCGAAUUGUUUCAG--CGCCGCUCGGCAGUUGUCCACAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCCCUUU----- ...(..((..((((.((((..(((--((((....))).))))...)))).))))..........(((((((....)))))))))..).......----- ( -26.40, z-score = -2.62, R) >droYak2.chr2R 20688381 92 - 21139217 CUUGUGGUAAUCGAAUUGUUUCAG--CGCCGCUCGGCAGUUGUCCACAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCCCUUU----- ...(..((..((((.((((..(((--((((....))).))))...)))).))))..........(((((((....)))))))))..).......----- ( -26.40, z-score = -2.62, R) >droEre2.scaffold_4845 22092002 92 - 22589142 CUUGUGGUAAUCGAAUUGUUUCAG--CGCCGCUCGGCAGUUGUCCACAAACCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCCCUUU----- .((((((((((.(((....)))..--.(((....))).)))).))))))...(((..(((....(((((((....)))))))...)))..))).----- ( -25.20, z-score = -2.06, R) >droAna3.scaffold_13266 12013407 97 - 19884421 CUCGUGGUAAUCGAAUUGUUUCAG--CGCCGCUCGGCAGUUGUCCGCAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCCGCUCCGUUCCCUUU ...(((((..((((.((((..(((--((((....))).))))...)))).))))..........(((((((....))))))))))))............ ( -32.50, z-score = -3.86, R) >dp4.chr3 8993621 93 + 19779522 UUUGUGGUAAUCGAAUUGUUUCAG--CACCGCUCGGCAGUUGUCCGCAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCUCUUUA---- (((((((....(.(((((((..((--(...))).)))))))).)))))))..(((..(((....(((((((....)))))))...))))))....---- ( -23.30, z-score = -1.53, R) >droPer1.super_4 1053578 93 + 7162766 UUUGUGGUAAUCGAAUUGUUUCAG--CGCCGCUCGGCAGUUGUCCGCAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCUCUUUA---- ...(..((..((((.((((..(((--((((....))).))))...)))).))))..........(((((((....)))))))))..)........---- ( -26.70, z-score = -2.37, R) >droVir3.scaffold_12875 1473847 87 - 20611582 -------UAAUCGAAUUGUUUCAGAGCGCCGCUCGACGGCCGUCCGCAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGUCGUUAUUGUU----- -------...((((.((((....((((...))))(((....))).)))).))))..((((....(((((((....)))))))......))))..----- ( -23.40, z-score = -1.65, R) >droMoj3.scaffold_6496 4578774 74 - 26866924 -------UAAUCGAAUUGUUUCAG-------------CGCCGUCCGCAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGUCAUUAAUGUU----- -------.....((((((.(((.(-------------((.....))).....))).))))))..(((((((....)))))))............----- ( -14.80, z-score = -1.82, R) >droGri2.scaffold_15245 8803343 85 + 18325388 -------UAAUCGAAUUGUUUCAG--CGCCGCUCGACAGUCGUCCGCAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGUUAUUAUUGUU----- -------...((((.((((....(--((.(........).)))..)))).))))..((((....(((((((....)))))))......))))..----- ( -18.60, z-score = -1.52, R) >consensus CUUGUGGUAAUCGAAUUGUUUCAG__CGCCGCUCGGCAGUUGUCCGCAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGUCAAGCUGUUCCCUUU_____ ............((((((............((((((..((.....))...))))))))))))..(((((((....)))))))................. (-15.01 = -14.95 + -0.06)



| Location | 20,727,325 – 20,727,422 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.69 |
| Shannon entropy | 0.31096 |
| G+C content | 0.49592 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.91 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20727325 97 - 21146708 ------UCAGUGGUUGUU--GCCCACCGCUUGUGGUAAUCGAAUUGUUUCAGCGCCGCUCGGCAGUUGUCCA-CAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGU ------...((((.....--..)))).((((.(((...((((.((((..(((((((....))).))))...)-))).))))((((((...))))))..))).)))) ( -30.00, z-score = -2.15, R) >droSim1.chr2R 19175399 97 - 19596830 ------CCAGUGGUUGUU--GCCCACCGCUUGUGGUAAUCGAAUUGUUUCAGCGCCGCUCGGCAGUUGUCCA-CAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGU ------...((((.....--..)))).((((.(((...((((.((((..(((((((....))).))))...)-))).))))((((((...))))))..))).)))) ( -30.00, z-score = -2.13, R) >droSec1.super_23 583820 97 - 989336 ------CCAGUGGUUGUU--GCCCACCGCUUGUGGUAAUCGAAUUGUUUCAGCGCCGCUCGGCAGUUGUCCA-CAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGU ------...((((.....--..)))).((((.(((...((((.((((..(((((((....))).))))...)-))).))))((((((...))))))..))).)))) ( -30.00, z-score = -2.13, R) >droYak2.chr2R 20688396 82 - 21139217 -----------------------UACCGCUUGUGGUAAUCGAAUUGUUUCAGCGCCGCUCGGCAGUUGUCCA-CAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGU -----------------------....((((.(((...((((.((((..(((((((....))).))))...)-))).))))((((((...))))))..))).)))) ( -26.30, z-score = -3.05, R) >droEre2.scaffold_4845 22092017 97 - 22589142 ------CCAGUGGCUGUU--GCCUACCGCUUGUGGUAAUCGAAUUGUUUCAGCGCCGCUCGGCAGUUGUCCA-CAAACCGAGUCAAUUUAAUUGACCUCCAGAGGU ------...(((((....--))).)).((((.(((...(((..((((..(((((((....))).))))...)-)))..)))((((((...))))))..))).)))) ( -28.40, z-score = -1.20, R) >droAna3.scaffold_13266 12013427 94 - 19884421 --------CGUACUUGCG--ACCC-CCUCUCGUGGUAAUCGAAUUGUUUCAGCGCCGCUCGGCAGUUGUCCG-CAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGU --------(((....)))--....-(((((.(.(((..((((.((((..(((((((....))).))))...)-))).))))..((((...))))))).).))))). ( -27.70, z-score = -1.96, R) >dp4.chr3 8993637 96 + 19779522 ------GUAGUUGUUGCU--ACCC-CCUUUUGUGGUAAUCGAAUUGUUUCAGCACCGCUCGGCAGUUGUCCG-CAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGU ------(((((....)))--))..-.((((((..(...((((.((((..((((.((....))..))))...)-))).))))((((((...)))))))..)))))). ( -25.20, z-score = -1.94, R) >droWil1.scaffold_180700 6428336 106 + 6630534 GUAGUUGUUGUUGUUGUUUGACCCGGUCCCCCACCUAAUCGAAUUGUUUCAGCGCCGCUCGACAAUUGUCCGCCAAAACGAGUCAAUUUAAUUGACCUCCAGAGGU ......((((..(..((((((...(((.....)))...))))))..)..))))(((((..(((....))).))......((((((((...))))).)))....))) ( -20.60, z-score = -0.17, R) >consensus ______CCAGUGGUUGUU__GCCCACCGCUUGUGGUAAUCGAAUUGUUUCAGCGCCGCUCGGCAGUUGUCCA_CAAAUCGAGUCAAUUUAAUUGACCUCCAGAGGU ............................(((.(((...((((.(((...(((((((....))).)))).....))).))))((((((...))))))..))).))). (-16.88 = -17.91 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:14 2011