| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,630,142 – 20,630,216 |
| Length | 74 |
| Max. P | 0.902847 |

| Location | 20,630,142 – 20,630,216 |
|---|---|
| Length | 74 |
| Sequences | 12 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 92.93 |
| Shannon entropy | 0.15413 |
| G+C content | 0.33435 |
| Mean single sequence MFE | -15.62 |
| Consensus MFE | -13.22 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.902847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
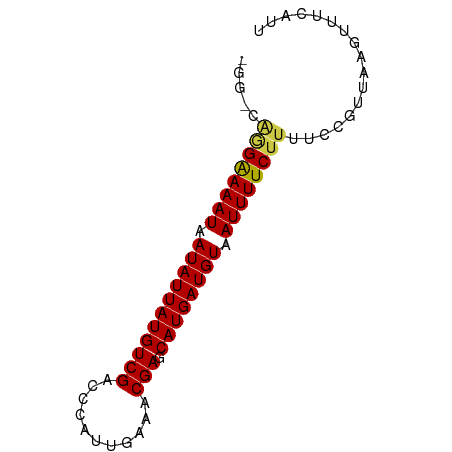
>dm3.chr2R 20630142 74 - 21146708 GGGGCAGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU .(((.((((((((.(((((((((((...........))).)))))))).)))))))).)))............. ( -18.70, z-score = -2.41, R) >droSim1.chr2R 19078676 74 - 19596830 GGGGCAGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU .(((.((((((((.(((((((((((...........))).)))))))).)))))))).)))............. ( -18.70, z-score = -2.41, R) >droSec1.super_23 488951 74 - 989336 GGGGCAGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU .(((.((((((((.(((((((((((...........))).)))))))).)))))))).)))............. ( -18.70, z-score = -2.41, R) >droYak2.chr2R 20591256 74 - 21139217 GAGCCAGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU (((..((((((((.(((((((((((...........))).)))))))).))))))))))).............. ( -13.90, z-score = -1.72, R) >droEre2.scaffold_4845 21994066 74 - 22589142 GGGCCAGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU .((..((((((((.(((((((((((...........))).)))))))).))))))))..))............. ( -17.00, z-score = -2.03, R) >droAna3.scaffold_13266 7215209 74 - 19884421 GGGCCGAGAAAAUAAUAUUAUGUCGACCCAUUGAGACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU .((..((((((((.(((((((((((...........))).)))))))).))))))))..))............. ( -17.70, z-score = -2.23, R) >dp4.chr3 3977666 67 + 19779522 -------GGAAAUAAUAUUAUGUCGACCCAUUGAGACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU -------((((((.(((((((((((...........))).)))))))).))))))................... ( -11.90, z-score = -1.18, R) >droPer1.super_4 960612 67 - 7162766 -------GGAAAUAAUAUUAUGUCGACCCAUUGAGACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU -------((((((.(((((((((((...........))).)))))))).))))))................... ( -11.90, z-score = -1.18, R) >droWil1.scaffold_180700 2317975 73 + 6630534 -GAUGAAGAAAAUAAUAUUAUGUCGACCCAUUGAGACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCACU -(..(((((((((.(((((((((((...........))).)))))))).)))))))))..)............. ( -16.20, z-score = -2.73, R) >droMoj3.scaffold_6496 4203314 71 - 26866924 ---GAAGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU ---((((((((((.(((((((((((...........))).)))))))).))))))))))............... ( -15.70, z-score = -2.64, R) >droVir3.scaffold_12875 1136644 69 - 20611582 -----AGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU -----((((((((.(((((((((((...........))).)))))))).))))))))................. ( -13.50, z-score = -1.99, R) >droGri2.scaffold_15245 8482749 69 + 18325388 -----AGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU -----((((((((.(((((((((((...........))).)))))))).))))))))................. ( -13.50, z-score = -1.99, R) >consensus _GG_CAGGAAAAUAAUAUUAUGUCGACCCAUUGAAACGAGCAUGAUGUAAUUUUCUUUUCCGUUAAGUUUCAUU .....((((((((.(((((((((((...........))).)))))))).))))))))................. (-13.22 = -13.24 + 0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:08 2011